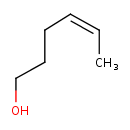
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-056u-9000000000-028a8b3df4d4f239877f | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9200000000-e53b385c79c9575f78e6 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-9600000000-a087b7d2729a1b64b371 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9200000000-ae453da156ca6e02cd43 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-260d12673632f88849b5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-04ee26e1d76a08ecd25b | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9000000000-f23cfb6a1c515c38983d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00l7-9000000000-2f32e5018a5a0ae6c3b8 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014l-9000000000-e0b8b7abee28acec2f3f | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052f-9000000000-4d8509dcdd5c8286d795 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4u-9000000000-3ac7fd8a059613793260 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-658fe53778817eb82301 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001j-9000000000-8876baa48206a003e28c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aor-9000000000-69d4385fb1a0066b87d1 | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |