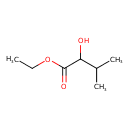
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00di-9000000000-758b88f178881da1325a | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0002-9700000000-827896d9063f78053cdf | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-2900000000-fe0008f310bae14bf859 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0005-9200000000-cef4feb543b20bbb416d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dl-9000000000-c600deb5779a6c477b93 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9700000000-f41001d0fa01ddfa33e9 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9200000000-593f7d39a8eb61924077 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fs-9000000000-457416d5b64dc2dfcf4c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-2900000000-0e3e2fc93ced4eaad704 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0005-9000000000-90eca02bf6ee993a042e | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abd-9000000000-8d9bdfd791bcedb98229 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-9300000000-8e1d59860df1f3587b7d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-9000000000-c77669f11b917e255d11 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-174016be00a6ebc6182e | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |