| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 22:40:35 UTC |
|---|
| Update Date | 2016-11-09 01:18:10 UTC |
|---|
| Accession Number | CHEM025912 |
|---|
| Identification |
|---|
| Common Name | Soyasapogenol B-1 |
|---|
| Class | Small Molecule |
|---|
| Description | A pentacyclic triterpenoid that is oleanane containing a double bond between positions 12 and 13 and substituted by hydroxy groups at the 3beta, 22beta and 24-positions. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
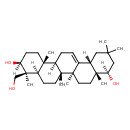
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 24-Hydroxysophoradiol | ChEBI | | Soyasapogenol-b | ChEBI | | (3beta,4beta,22beta)-Olean-12-ene-3,22,23-triol | HMDB | | (3Β,4β,22β)-olean-12-ene-3,22,23-triol | HMDB | | 3beta,22beta,24-Trihydroxyolean-12-ene | HMDB | | 3Β,22β,24-trihydroxyolean-12-ene | HMDB | | Olean-12-en-3beta,22beta,24-triol | HMDB | | Olean-12-en-3β,22β,24-triol | HMDB | | Olean-12-ene-3beta,22beta,24-triol | HMDB | | Olean-12-ene-3β,22β,24-triol | HMDB | | Soyasapogenin b | HMDB | | Soyasapogenol I | HMDB | | Soyasapogenol B | HMDB |
|
|---|
| Chemical Formula | C30H50O3 |
|---|
| Average Molecular Mass | 458.727 g/mol |
|---|
| Monoisotopic Mass | 458.376 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | (3S,4S,4aR,6aR,6bS,8aR,9R,12aS,14aR,14bR)-4-(hydroxymethyl)-4,6a,6b,8a,11,11,14b-heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,14,14a,14b-icosahydropicene-3,9-diol |
|---|
| Traditional Name | soyasapogenol B |
|---|
| SMILES | [H][C@]1(O)CC[C@@]2(C)[C@@]([H])(CC[C@]3(C)[C@]2([H])CC=C2[C@]4([H])CC(C)(C)C[C@@]([H])(O)[C@]4(C)CC[C@@]32C)[C@@]1(C)CO |
|---|
| InChI Identifier | InChI=1S/C30H50O3/c1-25(2)16-20-19-8-9-22-27(4)12-11-23(32)28(5,18-31)21(27)10-13-30(22,7)29(19,6)15-14-26(20,3)24(33)17-25/h8,20-24,31-33H,9-18H2,1-7H3/t20-,21+,22+,23-,24+,26+,27-,28+,29+,30+/m0/s1 |
|---|
| InChI Key | YOQAQNKGFOLRGT-UXXABWCISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Steroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-0000900000-2fc48e21e55611b63ab6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0021900000-52addc93ebec64414d6a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0v4l-1497400000-c27de81ed79f40b73292 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-01030582404004ad5282 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-0000900000-a318ff663fac4b5bb7a4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-08i0-0001900000-9b953b18916be4d05d57 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-2e8eabc98b7e3f409adf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0000900000-f4d5ff942354965a0479 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0000900000-85f3e3f9f2c6c002e35c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0abc-0000900000-83b5238e3b1963b51472 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0322900000-7cdf6e41b37aaae050e8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-2973000000-6f93c5f08d55f48fabac | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0034648 |
|---|
| FooDB ID | FDB005622 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000256 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | CPD-7610 |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 102937 |
|---|
| ChEBI ID | 9209 |
|---|
| PubChem Compound ID | 115012 |
|---|
| Kegg Compound ID | C08980 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|