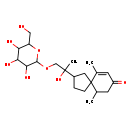| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 22:00:42 UTC |
|---|
| Update Date | 2016-11-09 01:17:58 UTC |
|---|
| Accession Number | CHEM024919 |
|---|
| Identification |
|---|
| Common Name | (4R,5S,7R,11x)-11,12-Dihydroxy-1(10)-spirovetiven-2-one 12-glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | (4R,5S,7R,11x)-11,12-Dihydroxy-1(10)-spirovetiven-2-one 12-glucoside is found in potato. (4R,5S,7R,11x)-11,12-Dihydroxy-1(10)-spirovetiven-2-one 12-glucoside is isolated from potatoes infected with Phoma exigu |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C21H34O8 |
|---|
| Average Molecular Mass | 414.490 g/mol |
|---|
| Monoisotopic Mass | 414.225 g/mol |
|---|
| CAS Registry Number | 62574-29-2 |
|---|
| IUPAC Name | 2-(2-hydroxy-1-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}propan-2-yl)-6,10-dimethylspiro[4.5]dec-6-en-8-one |
|---|
| Traditional Name | 2-(2-hydroxy-1-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}propan-2-yl)-6,10-dimethylspiro[4.5]dec-6-en-8-one |
|---|
| SMILES | CC1CC(=O)C=C(C)C11CCC(C1)C(C)(O)COC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C21H34O8/c1-11-6-14(23)7-12(2)21(11)5-4-13(8-21)20(3,27)10-28-19-18(26)17(25)16(24)15(9-22)29-19/h6,12-13,15-19,22,24-27H,4-5,7-10H2,1-3H3 |
|---|
| InChI Key | FSQYQQPZIHCQMQ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as terpene glycosides. These are prenol lipids containing a carbohydrate moiety glycosidically bound to a terpene backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Terpene glycosides |
|---|
| Direct Parent | Terpene glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Terpene glycoside
- Spirovetivane-type sesquiterpenoid
- Sesquiterpenoid
- Hexose monosaccharide
- Glycosyl compound
- O-glycosyl compound
- Cyclohexenone
- Monosaccharide
- Oxane
- Tertiary alcohol
- Ketone
- Cyclic ketone
- Secondary alcohol
- Acetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Organooxygen compound
- Primary alcohol
- Carbonyl group
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000b-7928000000-8469cc4f98249e86d40e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-014i-3283219000-a2b7a5e87699d68ce345 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0479700000-31e51cf560af7bef464c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fbj-1893000000-3b0112d3c9284acfa155 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uy0-1941000000-6a2843fa3f1133063430 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3653900000-8257da0f64305318f269 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-4942100000-0800ae9b0fcd631ffc91 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ki6-9340000000-fedeb50b72a59c91e04b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000900000-5bc0ddb692c019f1d5f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-8297800000-17ca89258a1fa6723989 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0btc-9141200000-f94a284465eba3a95eaa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gbj-0297700000-0da0427350c02b29700f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-2953000000-f2ad81b7b8b937ce7220 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gba-3910000000-63c8f9b3ebfb2bae12f6 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030895 |
|---|
| FooDB ID | FDB002859 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 74886379 |
|---|
| ChEBI ID | 168137 |
|---|
| PubChem Compound ID | 131751091 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|