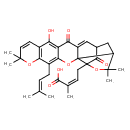| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:53:56 UTC |
|---|
| Update Date | 2016-11-09 01:17:57 UTC |
|---|
| Accession Number | CHEM024746 |
|---|
| Identification |
|---|
| Common Name | Isomorellic acid |
|---|
| Class | Small Molecule |
|---|
| Description | Morellic acid is found in fruits. Morellic acid is from Garcinia morella (batuan |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Isomorellate | Generator | | Butyl hexanoic acid | HMDB | | Butyl caproate | HMDB | | Butyl caprylate | HMDB | | Butyl ester OF hexanoic acid | HMDB | | FEMA 2201 | HMDB | | Hexanoic acid, butyl ester | HMDB | | N-Butyl caproate | HMDB | | N-Butyl hexanoate | HMDB | | N-Butyl N-hexanoate | HMDB | | N-Caproic acid N-butyl ester | HMDB | | (2Z)-4-[12-Hydroxy-8,8,21,21-tetramethyl-5-(3-methylbut-2-en-1-yl)-14,18-dioxo-3,7,20-trioxahexacyclo[15.4.1.0²,¹⁵.0²,¹⁹.0⁴,¹³.0⁶,¹¹]docosa-4(13),5,9,11,15-pentaen-19-yl]-2-methylbut-2-enoate | HMDB | | Morellic acid | HMDB |
|
|---|
| Chemical Formula | C33H36O8 |
|---|
| Average Molecular Mass | 560.634 g/mol |
|---|
| Monoisotopic Mass | 560.241 g/mol |
|---|
| CAS Registry Number | 5304-71-2 |
|---|
| IUPAC Name | (2Z)-4-[12-hydroxy-8,8,21,21-tetramethyl-5-(3-methylbut-2-en-1-yl)-14,18-dioxo-3,7,20-trioxahexacyclo[15.4.1.0²,¹⁵.0²,¹⁹.0⁴,¹³.0⁶,¹¹]docosa-4,6(11),9,12,15-pentaen-19-yl]-2-methylbut-2-enoic acid |
|---|
| Traditional Name | (2Z)-4-[12-hydroxy-8,8,21,21-tetramethyl-5-(3-methylbut-2-en-1-yl)-14,18-dioxo-3,7,20-trioxahexacyclo[15.4.1.0²,¹⁵.0²,¹⁹.0⁴,¹³.0⁶,¹¹]docosa-4,6(11),9,12,15-pentaen-19-yl]-2-methylbut-2-enoic acid |
|---|
| SMILES | CC(C)=CCC1=C2OC34C5CC(C=C3C(=O)C2=C(O)C2=C1OC(C)(C)C=C2)C(=O)C4(C\C=C(\C)C(O)=O)OC5(C)C |
|---|
| InChI Identifier | InChI=1S/C33H36O8/c1-16(2)8-9-20-26-19(11-12-30(4,5)39-26)24(34)23-25(35)21-14-18-15-22-31(6,7)41-32(28(18)36,13-10-17(3)29(37)38)33(21,22)40-27(20)23/h8,10-12,14,18,22,34H,9,13,15H2,1-7H3,(H,37,38)/b17-10- |
|---|
| InChI Key | COVMVPHACFXMAX-YVLHZVERSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyranoxanthones. These are organic aromatic compounds containing a pyran or a hydrogenated derivative fused to a xanthone ring system. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Benzopyrans |
|---|
| Sub Class | 1-benzopyrans |
|---|
| Direct Parent | Pyranoxanthones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyranoxanthone
- Pyranochromene
- 2,2-dimethyl-1-benzopyran
- Chromone
- Aromatic monoterpenoid
- Monoterpenoid
- Aryl ketone
- Alkyl aryl ether
- Cyclohexenone
- Oxepane
- Benzenoid
- Vinylogous acid
- Tetrahydrofuran
- Ketone
- Monocarboxylic acid or derivatives
- Oxacycle
- Ether
- Dialkyl ether
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-3000190000-5ccd58feb8356dce5577 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-014i-4100039000-4e27ff9691f49b43df0c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Isomorellic acid,1TMS,#1" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03xu-0010190000-e6580faa402eb6f281a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014r-5020890000-b0762af03545984977bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ap0-3090500000-a91493ab4b0719665376 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0010190000-f1a314f26866136b6250 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0aor-1032290000-90525eaf06c7235ee276 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05g3-2691120000-1ca2694685abe5c7ebcb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000090000-e1ff69a699d97b46410f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-08fu-0000290000-1763d257815dd698f7c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6x-6000980000-f0ed7b52eb2fd654f7e5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000090000-47ed5b46695395a54cfb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0000190000-2a63176a1c63f8d1ed28 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05mo-3080930000-c3e5e4499c90419fed9b | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030734 |
|---|
| FooDB ID | FDB002660 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 11791 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 12294 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|