| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:41:48 UTC |
|---|
| Update Date | 2016-11-09 01:17:53 UTC |
|---|
| Accession Number | CHEM024434 |
|---|
| Identification |
|---|
| Common Name | 2-Amino-3,4-dihydroxypentanedioic acid |
|---|
| Class | Small Molecule |
|---|
| Description | 2-Amino-3,4-dihydroxypentanedioic acid is found in brassicas. 2-Amino-3,4-dihydroxypentanedioic acid is isolated from Lepidium sativum (garden cress) and Rheum rhaponticum (rhubarb |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
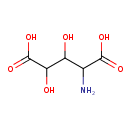
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Amino-3,4-dihydroxypentanedioate | Generator | | 3,4-Dihydroxyglutamic acid | HMDB | | (3S,4S)-DHGA | HMDB | | 3,4-Dihydroxy-L-glutamic acid | HMDB |
|
|---|
| Chemical Formula | C5H9NO6 |
|---|
| Average Molecular Mass | 179.128 g/mol |
|---|
| Monoisotopic Mass | 179.043 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 2-amino-3,4-dihydroxypentanedioic acid |
|---|
| Traditional Name | 2-amino-3,4-dihydroxypentanedioic acid |
|---|
| SMILES | NC(C(O)C(O)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H9NO6/c6-1(4(9)10)2(7)3(8)5(11)12/h1-3,7-8H,6H2,(H,9,10)(H,11,12) |
|---|
| InChI Key | SODSBJHVKSGOIZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamic acid or derivatives
- Alpha-amino acid
- Amino fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Short-chain hydroxy acid
- Alpha-hydroxy acid
- Dicarboxylic acid or derivatives
- Hydroxy acid
- Monosaccharide
- Fatty acid
- Fatty acyl
- 1,3-aminoalcohol
- 1,2-diol
- Amino acid
- Secondary alcohol
- Carboxylic acid
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Primary aliphatic amine
- Carbonyl group
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Primary amine
- Organonitrogen compound
- Organic nitrogen compound
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00fu-9200000000-e6bbcc0236ddba145fbd | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0udi-2119100000-ef888eb76542c79ee998 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-1900000000-ce17914d1c6b95adb671 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00xr-9600000000-350435acb4ecc7c19049 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-9200000000-3f3415eabdb4ad4cfa72 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0fai-4900000000-28171dc5948decfd9e28 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-9700000000-095a92f4556bf1ec61eb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9100000000-039e4cc28a473d0a5548 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001l-1900000000-2fdcfd732a371240941d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-6900000000-cdbe9fab926c266b8a8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fr-9000000000-d00dfbb6acb27942df59 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0umi-5900000000-3a78787e24927a52753e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fk9-9400000000-2cedb51d29f25218de90 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-878d6f90d5a2a6ef0fd6 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030408 |
|---|
| FooDB ID | FDB002269 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 11345033 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 22326306 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|