| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:13:04 UTC |
|---|
| Update Date | 2016-11-09 01:17:45 UTC |
|---|
| Accession Number | CHEM023664 |
|---|
| Identification |
|---|
| Common Name | Smilagenin |
|---|
| Class | Small Molecule |
|---|
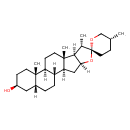
| Description | An oxaspiro compound that is(5beta,25R)-spirostan substituted by a beta-hydroxy group at position 3. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Isosarsapogenin | ChEBI | | (25R)-5beta-Spirostan-3beta-ol | Kegg | | (25R)-5b-Spirostan-3b-ol | Generator | | (25R)-5Β-spirostan-3β-ol | Generator | | Epi-sarsasapogenin | MeSH | | Epismilagenin | MeSH | | Sarsaponin | MeSH | | Sarsasapogenin | MeSH | | Sarsasapogenin, (3beta,5alpha,25R)-isomer | MeSH | | Sarsasapogenin, (3beta,5alpha,25S)-isomer | MeSH | | Sarsasapogenin, (3beta,5beta)-isomer | MeSH | | Sarsasapogenin, (3beta,5beta,25R)-isomer | MeSH | | Sarsasapogenin, (3beta,5beta,25S)-isomer | MeSH | | Tigogenin | MeSH | | (25R)-Spirostan-3beta-ol | HMDB | | 5beta -Spirostan-3beta (25R)--ol | HMDB | | 5beta-Spirostan-3beta-ol, (25R)- (8ci) | HMDB | | Isosarsasapogenin | HMDB | | Spirostan-3-ol, (3beta,5beta,25R)- (9ci) | HMDB | | Smilagenin | ChEBI |
|
|---|
| Chemical Formula | C27H44O3 |
|---|
| Average Molecular Mass | 416.637 g/mol |
|---|
| Monoisotopic Mass | 416.329 g/mol |
|---|
| CAS Registry Number | 126-18-1 |
|---|
| IUPAC Name | (1'R,2R,2'S,4'S,5R,7'S,8'R,9'S,12'S,13'S,16'S,18'R)-5,7',9',13'-tetramethyl-5'-oxaspiro[oxane-2,6'-pentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icosane]-16'-ol |
|---|
| Traditional Name | smilagenin |
|---|
| SMILES | [H][C@]12C[C@@]3([H])[C@]4([H])CC[C@]5([H])C[C@@H](O)CC[C@]5(C)[C@@]4([H])CC[C@]3(C)[C@@]1([H])[C@H](C)[C@@]1(CC[C@@H](C)CO1)O2 |
|---|
| InChI Identifier | InChI=1S/C27H44O3/c1-16-7-12-27(29-15-16)17(2)24-23(30-27)14-22-20-6-5-18-13-19(28)8-10-25(18,3)21(20)9-11-26(22,24)4/h16-24,28H,5-15H2,1-4H3/t16-,17+,18-,19+,20-,21+,22+,23+,24+,25+,26+,27-/m1/s1 |
|---|
| InChI Key | GMBQZIIUCVWOCD-UQHLGXRBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triterpenoids. These are terpene molecules containing six isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Triterpenoids |
|---|
| Direct Parent | Triterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Spirostane skeleton
- 3-hydroxysteroid
- Hydroxysteroid
- 3-beta-hydroxysteroid
- Steroid
- Ketal
- Oxane
- Tetrahydrofuran
- Cyclic alcohol
- Secondary alcohol
- Acetal
- Organoheterocyclic compound
- Oxacycle
- Alcohol
- Organic oxygen compound
- Organooxygen compound
- Hydrocarbon derivative
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pbi-2129100000-ce00917c9402bf19f2cb | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fr-5206900000-6994401f8cd0564b077a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-5039500000-2dc535b5234668294429 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0avi-5094100000-73cb8de52ed2c81a6008 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pvi-9055000000-cd920b055b8c4a5478c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-5003900000-c46f2a81526a3824ec01 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014j-2019200000-3a3da9fcf5a22a3b2993 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gb9-9025000000-662616db765aefc27d75 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-67ea14fb7a26b9b2d929 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0001900000-ff4f5d155a47a38a5c07 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02ta-0229700000-519f15705a81d147ae88 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0043900000-2b302fdea4f63c5306c1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ar0-1193300000-17ee1df0d3203669d2c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00b9-2590000000-9a82cac1ff37947e3a4a | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030048 |
|---|
| FooDB ID | FDB001354 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00003592 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 82568 |
|---|
| ChEBI ID | 28933 |
|---|
| PubChem Compound ID | 91439 |
|---|
| Kegg Compound ID | C08913 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|