| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:11:38 UTC |
|---|
| Update Date | 2016-11-09 01:17:45 UTC |
|---|
| Accession Number | CHEM023630 |
|---|
| Identification |
|---|
| Common Name | Scutigeral |
|---|
| Class | Small Molecule |
|---|
| Description | Scutigeral is found in mushrooms. Scutigeral is a constituent of Albatrellus ovinus |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
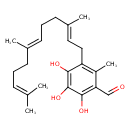
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Formyl-3-hydroxyneogrifolin | HMDB | | 2,3,4-Trihydroxy-6-methyl-5-(3,7,11-trimethyl-2,6,10-dodecatrienyl)benzaldehyde | HMDB | | Scutigeral | MeSH |
|
|---|
| Chemical Formula | C23H32O4 |
|---|
| Average Molecular Mass | 372.498 g/mol |
|---|
| Monoisotopic Mass | 372.230 g/mol |
|---|
| CAS Registry Number | 65195-50-8 |
|---|
| IUPAC Name | 2,3,4-trihydroxy-6-methyl-5-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]benzaldehyde |
|---|
| Traditional Name | 2,3,4-trihydroxy-6-methyl-5-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]benzaldehyde |
|---|
| SMILES | CC(C)=CCC\C(C)=C\CC\C(C)=C\CC1=C(C)C(C=O)=C(O)C(O)=C1O |
|---|
| InChI Identifier | InChI=1S/C23H32O4/c1-15(2)8-6-9-16(3)10-7-11-17(4)12-13-19-18(5)20(14-24)22(26)23(27)21(19)25/h8,10,12,14,25-27H,6-7,9,11,13H2,1-5H3/b16-10+,17-12+ |
|---|
| InChI Key | XQTQSUUULVXJPG-JTCWOHKRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sesquiterpenoids. These are terpenes with three consecutive isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sesquiterpenoid
- Farsesane sesquiterpenoid
- Benzenetriol
- Pyrogallol derivative
- Hydroxybenzaldehyde
- Benzaldehyde
- Benzoyl
- M-cresol
- P-cresol
- Phenol
- Toluene
- Aryl-aldehyde
- Monocyclic benzene moiety
- Benzenoid
- Vinylogous acid
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Aldehyde
- Organic oxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052r-5987000000-d1b25cf27cc62499f7ba | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00di-2200190000-ba3614291279f8d2b38a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0319000000-5e902eb480e350b03512 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0awc-1912000000-0ceefa66c20cf6741338 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014l-9851000000-8baf52c3b528c311c4ec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0009000000-f284b2c736278f0ad47e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0009000000-3e283b88b207b83210dc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-7977000000-2d8771ab118a64a61224 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-2529000000-74422bbdd85cec4ebcbf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001r-4946000000-d8cea8d4d06254cc129e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05ur-5900000000-7a4871bcd46bc13f9f85 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0009000000-7821d317d0c3d377d735 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0419000000-1348a7a81a0316cd324d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00bi-0493000000-1dd1238d4a90951d06bb | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030012 |
|---|
| FooDB ID | FDB001303 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00023940 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4476250 |
|---|
| ChEBI ID | 174956 |
|---|
| PubChem Compound ID | 5317377 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|