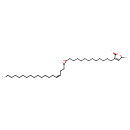
Epomusenin B (CHEM023602)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-05-25 21:10:33 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2016-11-09 01:17:44 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | CHEM023602 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Epomusenin B | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Epomusenin B is found in alcoholic beverages. Epomusenin B is a constituent of the fruit of Rollinia mucosa (biriba) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Contaminant Sources |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Contaminant Type | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C37H66O3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 558.918 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 558.501 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 178330-56-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 5-methyl-3-(12-{3-[(3Z)-octadec-3-en-1-yl]oxiran-2-yl}dodecyl)-2,5-dihydrofuran-2-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | 5-methyl-3-(12-{3-[(3Z)-octadec-3-en-1-yl]oxiran-2-yl}dodecyl)-5H-furan-2-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CCCCCCCCCCCCCC\C=C/CCC1OC1CCCCCCCCCCCCC1=CC(C)OC1=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C37H66O3/c1-3-4-5-6-7-8-9-10-11-12-13-14-18-21-24-27-30-35-36(40-35)31-28-25-22-19-16-15-17-20-23-26-29-34-32-33(2)39-37(34)38/h21,24,32-33,35-36H,3-20,22-23,25-31H2,1-2H3/b24-21- | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | MKKPDARISYEUBT-FLFQWRMESA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as annonaceous acetogenins. These are waxy derivatives of fatty acids (usually C32 or C34), containing a terminal carboxylic acid combined with a 2-propanol unit at the C-2 position to form a methyl- substituted alpha,beta-unsaturated-gamma-lactone. One of their interesting structural features is a single, adjacent, or nonadjacent tetrahydrofuran (THF) or tetrahydropyran (THP) system with one or two flanking hydroxyl group(s) at the center of a long hydrocarbon chain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty alcohols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Annonaceous acetogenins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0029983 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB001270 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | C00044157 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 8873424 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 10698082 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kegg Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| YMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ECMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||