| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:03:14 UTC |
|---|
| Update Date | 2016-11-09 01:17:42 UTC |
|---|
| Accession Number | CHEM023427 |
|---|
| Identification |
|---|
| Common Name | Protoisoeruboside B |
|---|
| Class | Small Molecule |
|---|
| Description | Protoisoeruboside B is found in onion-family vegetables. Protoisoeruboside B is a constituent of fresh garlic bulbs (Allium sativum) |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
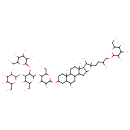
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 14-Pentyloxacyclotetradec-11-ene-2,11-dione | HMDB |
|
|---|
| Chemical Formula | C57H96O30 |
|---|
| Average Molecular Mass | 1261.354 g/mol |
|---|
| Monoisotopic Mass | 1260.599 g/mol |
|---|
| CAS Registry Number | 186545-34-6 |
|---|
| IUPAC Name | 2-({2-[(6-{[6,19-dihydroxy-7,9,13-trimethyl-6-(3-methyl-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}butyl)-5-oxapentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icosan-16-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl)oxy]-5-hydroxy-6-(hydroxymethyl)-3-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-4-yl}oxy)-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| Traditional Name | 2-({2-[(6-{[6,19-dihydroxy-7,9,13-trimethyl-6-(3-methyl-4-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}butyl)-5-oxapentacyclo[10.8.0.0²,⁹.0⁴,⁸.0¹³,¹⁸]icosan-16-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl)oxy]-5-hydroxy-6-(hydroxymethyl)-3-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-4-yl}oxy)-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| SMILES | CC(CCC1(O)OC2CC3C4CC(O)C5CC(CCC5(C)C4CCC3(C)C2C1C)OC1OC(CO)C(OC2OC(CO)C(O)C(OC3OC(CO)C(O)C(O)C3O)C2OC2OC(CO)C(O)C(O)C2O)C(O)C1O)COC1OC(CO)C(O)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C57H96O30/c1-20(19-77-50-43(72)39(68)35(64)29(14-58)79-50)5-10-57(76)21(2)34-28(87-57)13-25-23-12-27(63)26-11-22(6-8-55(26,3)24(23)7-9-56(25,34)4)78-51-46(75)42(71)47(33(18-62)83-51)84-54-49(86-53-45(74)41(70)37(66)31(16-60)81-53)48(38(67)32(17-61)82-54)85-52-44(73)40(69)36(65)30(15-59)80-52/h20-54,58-76H,5-19H2,1-4H3 |
|---|
| InChI Key | WYWLHHWQKOHXHW-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as steroidal saponins. These are saponins in which the aglycone moiety is a steroid. The steroidal aglycone is usually a spirostane, furostane, spirosolane, solanidane, or curcubitacin derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroidal glycosides |
|---|
| Direct Parent | Steroidal saponins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Steroidal saponin
- Diterpene glycoside
- Furostane-skeleton
- Oligosaccharide
- 22-hydroxysteroid
- Diterpenoid
- 6-hydroxysteroid
- Hydroxysteroid
- Terpene glycoside
- Fatty acyl glycoside
- Alkyl glycoside
- Glycosyl compound
- O-glycosyl compound
- Oxane
- Fatty acyl
- Cyclic alcohol
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Acetal
- Polyol
- Organoheterocyclic compound
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Primary alcohol
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001m-9060032402-0aba4951d8065b84d1c8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01qj-9140345607-50b2640a1246e67aeeb8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03gr-9451367508-bd406d1823c66c51b77d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002f-6390021202-f44535f4a2ac12e47826 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002f-6790023401-cea9e9d1d1b950317ed6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03fr-2901005101-b3b3a125a98c92ada242 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03ec-4090100000-f55d95a5ca99ce2fa7b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03em-6497350104-cc33d2dd788be2cf439d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06ri-6911102011-a12bd4a413863b0394a3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0190000000-6df97330b2b34ca4660b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-7970000000-09bb50264c2f2508f7ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abi-9311000101-f77cf2c2762e1ac99796 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0029785 |
|---|
| FooDB ID | FDB000992 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00057176 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 8058274 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 9882599 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|