| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 21:02:51 UTC |
|---|
| Update Date | 2016-11-09 01:17:42 UTC |
|---|
| Accession Number | CHEM023416 |
|---|
| Identification |
|---|
| Common Name | 6e,9e-Dihydroxy-4,7E-megastigmadien-3-one 9-[apiosyl-(1->6)-glucoside] |
|---|
| Class | Small Molecule |
|---|
| Description | 6S,9R-Dihydroxy-4,7E-megastigmadien-3-one 9-[apiosyl-(1->6)-glucoside] is found in fruits. 6S,9R-Dihydroxy-4,7E-megastigmadien-3-one 9-[apiosyl-(1->6)-glucoside] is isolated from Japanese medlar (Eriobotrya japonica) leaves. |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
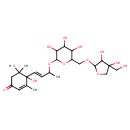
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C24H38O12 |
|---|
| Average Molecular Mass | 518.551 g/mol |
|---|
| Monoisotopic Mass | 518.236 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | 4-[(1E)-3-{[6-({[3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy}methyl)-3,4,5-trihydroxyoxan-2-yl]oxy}but-1-en-1-yl]-4-hydroxy-3,5,5-trimethylcyclohex-2-en-1-one |
|---|
| Traditional Name | 4-[(1E)-3-{[6-({[3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-yl]oxy}methyl)-3,4,5-trihydroxyoxan-2-yl]oxy}but-1-en-1-yl]-4-hydroxy-3,5,5-trimethylcyclohex-2-en-1-one |
|---|
| SMILES | CC(OC1OC(COC2OCC(O)(CO)C2O)C(O)C(O)C1O)\C=C\C1(O)C(C)=CC(=O)CC1(C)C |
|---|
| InChI Identifier | InChI=1S/C24H38O12/c1-12-7-14(26)8-22(3,4)24(12,32)6-5-13(2)35-20-18(29)17(28)16(27)15(36-20)9-33-21-19(30)23(31,10-25)11-34-21/h5-7,13,15-21,25,27-32H,8-11H2,1-4H3/b6-5+ |
|---|
| InChI Key | WGPMCTNBJPAHNW-AATRIKPKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Cyclofarsesane sesquiterpenoid
- Megastigmane sesquiterpenoid
- Sesquiterpenoid
- Ionone derivative
- Alkyl glycoside
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Cyclohexenone
- Oxane
- Tetrahydrofuran
- Tertiary alcohol
- Ketone
- Cyclic ketone
- Secondary alcohol
- Organoheterocyclic compound
- Acetal
- Oxacycle
- Polyol
- Alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-7592860000-d392b5e06307a150365a | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0592-9240007000-d9282fa7ac50eb7b2fa3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0kxr-1191150000-d6ecf1ec16dbaf49ba62 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-2391000000-4d1fe1632b5946caa429 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-6690000000-aa30fab3ffefdc2337cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01bj-0692540000-5fa7d3f657d27f809fa7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0691200000-99442a6cb501682da69a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-4890000000-f6e5ca40860e935b6ced | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0pbi-0974460000-0d31362add27692bcd5a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0aor-1693110000-039309a98922cb613266 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9362000000-89ceaa3cc4fbf7bc8780 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0110390000-d812abb514925595ae82 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0g4i-5913300000-0c6fe6d1e07644569a6a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pbc-7960000000-e2df906c346320f0f087 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0029773 |
|---|
| FooDB ID | FDB000980 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 168009 |
|---|
| PubChem Compound ID | 131750896 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|