| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 20:48:32 UTC |
|---|
| Update Date | 2016-11-09 01:17:37 UTC |
|---|
| Accession Number | CHEM023061 |
|---|
| Identification |
|---|
| Common Name | Gravacridonetriol glucoside |
|---|
| Class | Small Molecule |
|---|
| Description | Gravacridonetriol glucoside is found in herbs and spices. Gravacridonetriol glucoside is an alkaloid from Ruta graveolens (rue |
|---|
| Contaminant Sources | |
|---|
| Contaminant Type | Not Available |
|---|
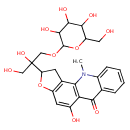
| Chemical Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C25H29NO11 |
|---|
| Average Molecular Mass | 519.498 g/mol |
|---|
| Monoisotopic Mass | 519.174 g/mol |
|---|
| CAS Registry Number | 59086-96-3 |
|---|
| IUPAC Name | 2-(1,2-dihydroxy-3-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}propan-2-yl)-5-hydroxy-11-methyl-1H,2H,6H,11H-furo[2,3-c]acridin-6-one |
|---|
| Traditional Name | 2-(1,2-dihydroxy-3-{[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}propan-2-yl)-5-hydroxy-11-methyl-1H,2H-furo[2,3-c]acridin-6-one |
|---|
| SMILES | CN1C2=CC=CC=C2C(=O)C2=C1C1=C(OC(C1)C(O)(CO)COC1OC(CO)C(O)C(O)C1O)C=C2O |
|---|
| InChI Identifier | InChI=1S/C25H29NO11/c1-26-13-5-3-2-4-11(13)20(30)18-14(29)7-15-12(19(18)26)6-17(36-15)25(34,9-28)10-35-24-23(33)22(32)21(31)16(8-27)37-24/h2-5,7,16-17,21-24,27-29,31-34H,6,8-10H2,1H3 |
|---|
| InChI Key | BFVREXFWNFMMSF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosylglycerols. These are glycerolipids structurally characterized by the presence of one or more sugar residues attached to glycerol via a glycosidic linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Glycosylglycerols |
|---|
| Direct Parent | Glycosylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycosylglycerol
- Acridone
- Acridine
- Benzoquinoline
- Hexose monosaccharide
- O-glycosyl compound
- Dihydroquinolone
- Glycosyl compound
- Quinoline
- Dihydroquinoline
- Coumaran
- 1-hydroxy-2-unsubstituted benzenoid
- Alkyl aryl ether
- Oxane
- Benzenoid
- Pyridine
- Monosaccharide
- Vinylogous amide
- Vinylogous acid
- Tertiary alcohol
- Heteroaromatic compound
- Secondary alcohol
- Acetal
- Ether
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Polyol
- Organic oxygen compound
- Hydrocarbon derivative
- Organic nitrogen compound
- Organic oxide
- Alcohol
- Organopnictogen compound
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zmr-9621850000-2158d6d8fffc0248f23f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0002-5401109000-21abfb91a119f1ad4a34 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0073190000-95a3f106fab3657255d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ktf-0369120000-d1628b06c5b1a0102c42 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000l-0090000000-83df6bab38a71aefa28d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-016r-2537590000-0b69ec7b240660a1d0c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03k9-4897630000-70f1420f476fb0d9df32 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0kfx-9471000000-e8e198c7fde1119ad6bb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0009020000-125dd9f65002cebd1dfc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0w9d-6906160000-2420559b83936a815181 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-07bg-9331100000-d13ce6402629f72465eb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000090000-80810b45a64e92ea92ca | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pvi-4911130000-3a2a2f0db4f486cc8252 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0rk9-3094010000-f401b9d9439ff731dcfc | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0029331 |
|---|
| FooDB ID | FDB000392 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 74886370 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 131750851 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|