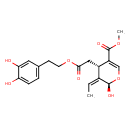| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 20:47:22 UTC |
|---|
| Update Date | 2016-11-09 01:17:37 UTC |
|---|
| Accession Number | CHEM023026 |
|---|
| Identification |
|---|
| Common Name | 3,4-DHPEA-EA |
|---|
| Class | Small Molecule |
|---|
| Description | 3,4-DHPEA-EA is found in olive. 3,4-DHPEA-EA is the major form of the oleuropein-aglycone |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,4-DHPEA-elenolic acid mono-aldehyde | HMDB | | Oleuropein-aglycone mono-aldehyde | HMDB | | Methyl (2R,4S)-4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-3-ethylidene-2-hydroxy-3,4-dihydro-2H-pyran-5-carboxylic acid | Generator | | 3,4-Dixydroxyphenylethanol elenolic acid | MeSH | | 3,4-DHPEA-ea | MeSH |
|
|---|
| Chemical Formula | C19H22O8 |
|---|
| Average Molecular Mass | 378.373 g/mol |
|---|
| Monoisotopic Mass | 378.131 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | methyl (2R,3Z,4S)-4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-3-ethylidene-2-hydroxy-3,4-dihydro-2H-pyran-5-carboxylate |
|---|
| Traditional Name | methyl (4S,5Z,6R)-4-{2-[2-(3,4-dihydroxyphenyl)ethoxy]-2-oxoethyl}-5-ethylidene-6-hydroxy-4,6-dihydropyran-3-carboxylate |
|---|
| SMILES | COC(=O)C1=CO[C@@H](O)\C(=C/C)[C@@H]1CC(=O)OCCC1=CC=C(O)C(O)=C1 |
|---|
| InChI Identifier | InChI=1S/C19H22O8/c1-3-12-13(14(18(23)25-2)10-27-19(12)24)9-17(22)26-7-6-11-4-5-15(20)16(21)8-11/h3-5,8,10,13,19-21,24H,6-7,9H2,1-2H3/b12-3-/t13-,19+/m0/s1 |
|---|
| InChI Key | BIWKXNFEOZXNLX-SQOYHTLWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as iridoids and derivatives. These are monoterpenes containing a skeleton structurally characterized by the presence of a cylopentane fused to a pyran ( forming a 4,7-dimethylcyclopenta[c]pyran), or a derivative where the pentane moiety is open. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Iridoids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Secoiridoid-skeleton
- Aromatic monoterpenoid
- Monocyclic monoterpenoid
- Tyrosol derivative
- Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- Phenol
- 1-hydroxy-2-unsubstituted benzenoid
- Monocyclic benzene moiety
- Benzenoid
- Dicarboxylic acid or derivatives
- Vinylogous ester
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Methyl ester
- Carboxylic acid ester
- Hemiacetal
- Oxacycle
- Carboxylic acid derivative
- Organoheterocyclic compound
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Organooxygen compound
- Organic oxygen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-3924000000-a774df901c6e416aa979 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0059-6040090000-2e8e4c02704c1060b458 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0769000000-44f59fc6a7f7eccf78b1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0922000000-cfa03854b1f3f2e1d692 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-6910000000-af8e65043b5cc4bd2027 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00b9-1549000000-0f8a81848af23892f824 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dm-1984000000-400bc643c703ff71eca9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abi-1900000000-af441007a9141cc14c96 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0129000000-3f25a8adebbfe0f30b8e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00vi-0926000000-196f5729bb68656d092a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-1911000000-aefd6a7daecc217d4d11 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03fr-0129000000-6f49095caa9d91c93b25 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0903000000-43a413396c8163d27ce7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01di-5902000000-66e3b9f0e854cf50afa2 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0029304 |
|---|
| FooDB ID | FDB000346 |
|---|
| Phenol Explorer ID | 688 |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 30776771 |
|---|
| ChEBI ID | 89421 |
|---|
| PubChem Compound ID | 56842347 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|