| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:55:51 UTC |
|---|
| Update Date | 2016-11-09 01:17:35 UTC |
|---|
| Accession Number | CHEM022833 |
|---|
| Identification |
|---|
| Common Name | (-)-beta-Phellandrene |
|---|
| Class | Small Molecule |
|---|
| Description | A beta-phellandrene in which the chiral centre has R configuration. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Feces
|
|---|
| Contaminant Type | Not Available |
|---|
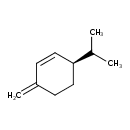
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-3-Methylene-6-(1-methylethyl)cyclohexene | ChEBI | | (-)-p-Mentha-1(7),2-diene | ChEBI | | (3R)-3-Isopropyl-6-methylenecyclohexene | ChEBI | | (R)-3-Isopropyl-6-methylenecyclohexene | ChEBI | | beta-Phellandrene L-form | ChEBI | | b-Phellandrene L-form | Generator | | Β-phellandrene L-form | Generator | | (-)-b-Phellandrene | Generator | | (-)-Β-phellandrene | Generator | | (6R)-3-Methylene-6-(1-methylethyl)cyclohexene | HMDB | | L-beta-Phellandrene | HMDB | | L-Β-phellandrene | HMDB | | p-Mentha-1(7),2-diene | HMDB | | 3-Methylene-6-(1-methylethyl)cyclohexene | HMDB | | (±)-beta-phellandrene | HMDB | | (±)-β-phellandrene | HMDB | | 3-Isopropyl-6-methylene-1-cyclohexene | HMDB | | 4-Isopropyl-1-methylene-2-cyclohexene | HMDB | | beta-Phellandrene | HMDB | | (-)-beta-Phellandrene | PhytoBank |
|
|---|
| Chemical Formula | C10H16 |
|---|
| Average Molecular Mass | 136.238 g/mol |
|---|
| Monoisotopic Mass | 136.125 g/mol |
|---|
| CAS Registry Number | 6153-17-9 |
|---|
| IUPAC Name | (6R)-3-methylidene-6-(propan-2-yl)cyclohex-1-ene |
|---|
| Traditional Name | (-)-β-phellandrene |
|---|
| SMILES | [H][C@@]1(CCC(=C)C=C1)C(C)C |
|---|
| InChI Identifier | InChI=1S/C10H16/c1-8(2)10-6-4-9(3)5-7-10/h4,6,8,10H,3,5,7H2,1-2H3/t10-/m1/s1 |
|---|
| InChI Key | LFJQCDVYDGGFCH-SNVBAGLBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as menthane monoterpenoids. These are monoterpenoids with a structure based on the o-, m-, or p-menthane backbone. P-menthane consists of the cyclohexane ring with a methyl group and a (2-methyl)-propyl group at the 1 and 4 ring position, respectively. The o- and m- menthanes are much rarer, and presumably arise by alkyl migration of p-menthanes. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Menthane monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - P-menthane monoterpenoid
- Monocyclic monoterpenoid
- Branched unsaturated hydrocarbon
- Cycloalkene
- Cyclic olefin
- Unsaturated aliphatic hydrocarbon
- Unsaturated hydrocarbon
- Olefin
- Hydrocarbon
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-2900000000-f7f288db31b30052203d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9600000000-48905d36448ddbdd3f79 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-1000-9100000000-344480f8a40ee3d9f098 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0900000000-2950ad058f77d7bc9f76 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0900000000-2146d0245e5c9841d77b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014r-7900000000-7c7a8d3355c299f4c547 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000j-9500000000-9054c1e940b7e2523fc6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000x-9100000000-2840ace0787e7be76083 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6x-9000000000-788626760da3cd443350 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0900000000-a013b4ae27f975ab5621 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0900000000-a013b4ae27f975ab5621 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9300000000-24aa02d1330c72611fdb | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0041633 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00010873 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | CPD-8768 |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 391433 |
|---|
| ChEBI ID | 129 |
|---|
| PubChem Compound ID | 443161 |
|---|
| Kegg Compound ID | C11392 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|