| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:55:10 UTC |
|---|
| Update Date | 2016-11-09 01:17:35 UTC |
|---|
| Accession Number | CHEM022821 |
|---|
| Identification |
|---|
| Common Name | Inositol phosphate |
|---|
| Class | Small Molecule |
|---|
| Description | A myo-inositol monophosphate in which the phosphate group is located at position 6. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Feces
|
|---|
| Contaminant Type | Not Available |
|---|
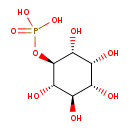
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| D-myo-Inositol (6)-monophosphate | ChEBI | | D-myo-Inositol (6)-phosphate | ChEBI | | I6P | ChEBI | | Ins(6)P | ChEBI | | Ins6P | ChEBI | | D-myo-Inositol (6)-monophosphoric acid | Generator | | Inositol phosphoric acid | Generator, HMDB | | D-myo-Inositol (6)-phosphoric acid | Generator | | 1D-myo-Inositol 6-phosphoric acid | Generator, HMDB | | D-myo-Inositol 6-phosphate | HMDB | | D-myo-Inositol-6-monophosphate | HMDB | | Inositol 1-phosphate | HMDB | | Inositol monophosphate | HMDB | | Inositol phosphate | HMDB | | Inositophosphoric acid | HMDB |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Mass | 260.136 g/mol |
|---|
| Monoisotopic Mass | 260.030 g/mol |
|---|
| CAS Registry Number | 15421-51-9 |
|---|
| IUPAC Name | {[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | [(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxyphosphonic acid |
|---|
| SMILES | O[C@@H]1[C@@H](O)[C@H](O)[C@@H](OP(O)(O)=O)[C@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-1-2(8)4(10)6(5(11)3(1)9)15-16(12,13)14/h1-11H,(H2,12,13,14)/t1-,2-,3-,4+,5-,6-/m1/s1 |
|---|
| InChI Key | INAPMGSXUVUWAF-XCMZKKERSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0005-9650000000-8feb65965bdf1456b458 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-0a4r-2184339000-409fe8e34ab4403363db | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_5_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("myo-Inositol 6-phosphate,5TBDMS,#5" TMS) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-2390000000-036684c0736cc37701a9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-2290000000-2692f0855292325fd753 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000w-9600000000-81c4994a2212052d4690 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-4190000000-f8cff124acad5d21332f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9220000000-2153b6c9b35a97307acb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-5f386828ffd5102695f3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-05d6010bb4bbf2b0c228 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6s-9280000000-69bd06f4ab8866ad9db1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-9000000000-f3e6675946c65f8f8f07 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0090000000-89fa9427c518aa95bbd0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0190000000-fa4c1ecfe995d290a153 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9100000000-fc4055901243469636ad | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0002985 |
|---|
| FooDB ID | FDB023090 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | 1484840 |
|---|
| BioCyc ID | CPD-6702 |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Inositol phosphate |
|---|
| Chemspider ID | 10465039 |
|---|
| ChEBI ID | 64838 |
|---|
| PubChem Compound ID | Not Available |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=1530577 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=17439666 | | 3. Pirrung, Michael C. Discovery of a catalytic asymmetric phosphorylation through selection of a minimal kinase mimic: A concise total synthesis of D-myo-inositol-1-phosphate. Chemtracts (2001), 14(14), 802-804. | | 4. Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. | | 5. Jooste EH, Sharma A, Zhang Y, Emala CW: Rapacuronium augments acetylcholine-induced bronchoconstriction via positive allosteric interactions at the M3 muscarinic receptor. Anesthesiology. 2005 Dec;103(6):1195-203. | | 6. Buccellati C, Sala A, Rossoni G, Capra V, Rovati GE, Di Gennaro A, Folco G, Colli S, Casagrande C: Pharmacological characterization of 2NTX-99 [4-methoxy-N1-(4-trans-nitrooxycyclohexyl)-N3-(3-pyridinylmethyl)-1,3-benzenedica rboxamide], a potential antiatherothrombotic agent with antithromboxane and nitric oxide donor activity in platelet and vascular preparations. J Pharmacol Exp Ther. 2006 May;317(2):830-7. Epub 2006 Jan 6. | | 7. Honchar MP, Olney JW, Sherman WR: Systemic cholinergic agents induce seizures and brain damage in lithium-treated rats. Science. 1983 Apr 15;220(4594):323-5. | | 8. Grimminger F, Rose F, Sibelius U, Meinhardt M, Potzsch B, Spriestersbach R, Bhakdi S, Suttorp N, Seeger W: Human endothelial cell activation and mediator release in response to the bacterial exotoxins Escherichia coli hemolysin and staphylococcal alpha-toxin. J Immunol. 1997 Aug 15;159(4):1909-16. | | 9. Kato T, Shioiri T, Takahashi S, Inubushi T: Measurement of brain phosphoinositide metabolism in bipolar patients using in vivo 31P-MRS. J Affect Disord. 1991 Aug;22(4):185-90. | | 10. Waldo GL, Corbitt J, Boyer JL, Ravi G, Kim HS, Ji XD, Lacy J, Jacobson KA, Harden TK: Quantitation of the P2Y(1) receptor with a high affinity radiolabeled antagonist. Mol Pharmacol. 2002 Nov;62(5):1249-57. | | 11. King WG, Rittenhouse SE: Inhibition of protein kinase C by staurosporine promotes elevated accumulations of inositol trisphosphates and tetrakisphosphate in human platelets exposed to thrombin. J Biol Chem. 1989 Apr 15;264(11):6070-4. | | 12. Chen WY, Ko FN, Lin CN, Teng CM: The effect of 3-[2-(cyclopropylamino)ethoxy]xanthone on platelet thromboxane formation. Thromb Res. 1994 Jul 1;75(1):81-90. | | 13. Siess W: Evidence for the formation of inositol 4-monophosphate in stimulated human platelets. FEBS Lett. 1985 Jun 3;185(1):151-6. | | 14. Fourcade O, Simon MF, Litt L, Samii K, Chap H: Propofol inhibits human platelet aggregation induced by proinflammatory lipid mediators. Anesth Analg. 2004 Aug;99(2):393-8, table of contents. | | 15. Ishii H, Umeda F, Hashimoto T, Nawata H: Increased inositol phosphate accumulation in platelets from patients with NIDDM. Diabetes Res Clin Pract. 1991 Oct;14(1):21-7. | | 16. Bae YS, Cantley LG, Chen CS, Kim SR, Kwon KS, Rhee SG: Activation of phospholipase C-gamma by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998 Feb 20;273(8):4465-9. | | 17. Williams RS: Pharmacogenetics in model systems: defining a common mechanism of action for mood stabilisers. Prog Neuropsychopharmacol Biol Psychiatry. 2005 Jul;29(6):1029-37. | | 18. Ross TS, Majerus PW: Inositol-1,2-cyclic-phosphate 2-inositolphosphohydrolase. Substrate specificity and regulation of activity by phospholipids, metal ion chelators, and inositol 2-phosphate. J Biol Chem. 1991 Jan 15;266(2):851-6. | | 19. Kories C, Czyborra C, Fetscher C, Schneider T, Krege S, Michel MC: Gender comparison of muscarinic receptor expression and function in rat and human urinary bladder: differential regulation of M2 and M3 receptors? Naunyn Schmiedebergs Arch Pharmacol. 2003 May;367(5):524-31. Epub 2003 Mar 28. | | 20. Morou E, Georgoussi Z: Expression of the third intracellular loop of the delta-opioid receptor inhibits signaling by opioid receptors and other G protein-coupled receptors. J Pharmacol Exp Ther. 2005 Dec;315(3):1368-79. Epub 2005 Sep 13. |
|
|---|