| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:50:59 UTC |
|---|
| Update Date | 2016-11-09 01:17:34 UTC |
|---|
| Accession Number | CHEM022756 |
|---|
| Identification |
|---|
| Common Name | Prolylhydroxyproline |
|---|
| Class | Small Molecule |
|---|
| Description | A dipeptide composed of L-proline and L-hydroxyproline residues. It is a biomarker for bone collagen degradation. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
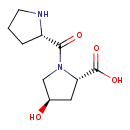
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Pro-L-hyp | ChEBI | | L-Prolyl-L-hydroxyproline | ChEBI | | (4R)-L-Prolyl-4-hydroxy-L-proline | HMDB | | 4-Hydroxy-1-L-prolyl-proline | HMDB | | L-4-Hydroxy-1-L-prolyl-proline | HMDB | | Proline-hydroxyproline | HMDB | | trans-4-Hydroxy-1-L-prolyl-proline | HMDB | | Prolylhydroxyproline | ChEBI | | Pro-hydroxy-Pro | HMDB | | Pro-Hyp | HMDB | | Proline hydroxyproline dipeptide | HMDB | | Proline-hydroxyproline dipeptide | HMDB |
|
|---|
| Chemical Formula | C10H16N2O4 |
|---|
| Average Molecular Mass | 228.245 g/mol |
|---|
| Monoisotopic Mass | 228.111 g/mol |
|---|
| CAS Registry Number | 18684-24-7 |
|---|
| IUPAC Name | (2S,4R)-4-hydroxy-1-[(2S)-pyrrolidine-2-carbonyl]pyrrolidine-2-carboxylic acid |
|---|
| Traditional Name | proline-hydroxyproline |
|---|
| SMILES | O[C@@H]1C[C@H](N(C1)C(=O)[C@@H]1CCCN1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H16N2O4/c13-6-4-8(10(15)16)12(5-6)9(14)7-2-1-3-11-7/h6-8,11,13H,1-5H2,(H,15,16)/t6-,7+,8+/m1/s1 |
|---|
| InChI Key | ONPXCLZMBSJLSP-CSMHCCOUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Proline or derivatives
- N-acyl-l-alpha-amino acid
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- N-acylpyrrolidine
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Pyrrolidine-2-carboxamide
- Tertiary carboxylic acid amide
- Pyrrolidine
- Secondary alcohol
- Amino acid
- Amino acid or derivatives
- Carboxamide group
- Secondary amine
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid
- Secondary aliphatic amine
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organopnictogen compound
- Organic oxide
- Alcohol
- Organonitrogen compound
- Carbonyl group
- Organooxygen compound
- Amine
- Organic nitrogen compound
- Organic oxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dj-9200000000-cefb6332c4f93d2283b3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0a4l-2692000000-7b1c4c7371558705bfb0 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-00di-9200000000-6af4dc9ae133d399503c | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0b90-2900000000-84bcb537803c3e383c98 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1490000000-d607eb5709327adc37c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0229-9520000000-53efc51e1a53a9bb992e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9200000000-c54979309cb483b47143 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-0690000000-385af9502303330863e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05o0-3920000000-648587bfd6a99e5a6a25 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0239-9200000000-e801efd4fdf93e14e8f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01u1-4890000000-317a4ceb981d4c1d3a2c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-2900000000-cb814d27ee8198848547 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-df69d58c4ec35c77f011 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01t9-5690000000-afc10370af26972ee32d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0229-9610000000-4039cba60bbe790e2e1c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9300000000-0dd3ad4a047284fe2d79 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0006695 |
|---|
| FooDB ID | FDB024028 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 10077215 |
|---|
| ChEBI ID | 74767 |
|---|
| PubChem Compound ID | 11902892 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=12636053 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=22382331 | | 3. ustova, V.; Blaha, Karel. Amino acids and peptides. CXXVI. Dipeptides containing proline and 4-hydroxyproline. Collection of Czechoslovak Chemical Communications (1975), 40(3), 662-9. | | 4. Pohlidal A, Husek P, Palicka V, Slabik D, Hill M, Matucha P: Novel and traditional biomarkers of bone turnover in postmenopausal women. Clin Chem Lab Med. 2003 Jan;41(1):74-8. | | 5. Husek P, Pohlidal A, Slabik D: Rapid screening of urinary proline-hydroxyproline dipeptide in bone turnover studies. J Chromatogr B Analyt Technol Biomed Life Sci. 2002 Feb 5;767(1):169-74. | | 6. Kodama H, Nakamura H, Sugahara K, Numajiri Y: Liquid chromatography-mass spectrometry for the qualitative analyses of iminodipeptides in the urine of patients with prolidase deficiency. J Chromatogr. 1990 May 18;527(2):279-88. | | 7. Sugahara K, Kodama H: Liquid chromatography-mass spectrometry for simultaneous analyses of iminodipeptides containing an N-terminal or a C-terminal proline. J Chromatogr. 1991 Apr 19;565(1-2):408-15. | | 8. Zhang J, Sugahara K, Yasuda K, Kodama H, Sagara Y, Kodama H: The effects of serum iminodipeptides and prednisolone on superoxide generation and tyrosyl phosphorylation of proteins in neutrophils from a patient with prolidase deficiency. Free Radic Biol Med. 1998 Mar 15;24(5):689-98. | | 9. Kawaguchi T, Nanbu PN, Kurokawa M: Distribution of prolylhydroxyproline and its metabolites after oral administration in rats. Biol Pharm Bull. 2012;35(3):422-7. | | 10. Lampiaho K, Nikkari T, Pikkarainen J, Karkkainen J, Kulonen E: Unexpected occurrence of prolylhydroxyproline during the analysis of collagen-bound carbohydrates by gas-liquid chromatography. J Chromatogr. 1972 Feb 2;64(2):211-8. | | 11. Bienenstock HKIBRICK AC: Urinary excretion of prolylhydroxyproline in rheumatic diseases. Ann Rheum Dis. 1969 Jan;28(1):28-30. | | 12. Codini M, Palmerini CA, Fini C, Lucarelli C, Floridi A: High-performance liquid chromatographic method for the determination of prolyl peptides in urine. J Chromatogr. 1991 Jan 4;536(1-2):337-41. | | 13. KIBRICK AC, HASHIRO CQ, SCHULTZ RS, WALTERS MI, MILHORAT AT: PROLYLHYDROXYPROLINE IN URINE: ITS DETERMINATION AND OBSERVATIONS IN MUSCULAR DYSTROPHY. Clin Chim Acta. 1964 Oct;10:344-51. | | 14. Hueckel HJ, Rogers QR: Prolylhydroxyproline absorption in hamsters. Can J Biochem. 1972 Jul;50(7):782-90. | | 15. Lowry M, Hall DE, Brosnan JT: Metabolism of glycine- and hydroxyproline-containing peptides by the isolated perfused rat kidney. Biochem J. 1985 Jul 15;229(2):545-9. | | 16. Kibrick AC, Power HL, Sevendal E, Milhorat AT: Prolylhydroxyproline in urine: improved method for detecting radioactivity with a scanner on full sheets of chromatography paper. Anal Biochem. 1968 Oct 24;25(1):40-5. | | 17. KIBRICK AC, HASHIRO CQ, WALTERS MI, MILHORAT AT: DIKETOPIPERAZINE OF PROLYLHYDROXYPROLINE IN NORMAL HUMAN URINE. Proc Soc Exp Biol Med. 1965 Jan;118:62-4. | | 18. Zhang A, Sun H, Han Y, Yuan Y, Wang P, Song G, Yuan X, Zhang M, Xie N, Wang X: Exploratory urinary metabolic biomarkers and pathways using UPLC-Q-TOF-HDMS coupled with pattern recognition approach. Analyst. 2012 Sep 21;137(18):4200-8. doi: 10.1039/c2an35780a. Epub 2012 Jul 31. | | 19. Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. |
|
|---|