| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:48:53 UTC |
|---|
| Update Date | 2016-11-09 01:17:34 UTC |
|---|
| Accession Number | CHEM022720 |
|---|
| Identification |
|---|
| Common Name | 6beta-Hydroxytestosterone |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
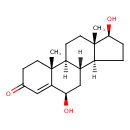
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Octadecanoyl-1-hexadecyl-sn-glycerol | HMDB | | (6beta,17beta)-6,17-Dihydroxyandrost-4-en-3-one | ChEBI | | 4-Androsten-6beta,17beta-diol-3-one | ChEBI | | 6beta,17beta-Dihydroxy-4-androsten-3-one | ChEBI | | 6beta,17beta-Dihydroxyandrost-4-en-3-one | ChEBI | | (6b,17b)-6,17-Dihydroxyandrost-4-en-3-one | Generator | | (6β,17β)-6,17-dihydroxyandrost-4-en-3-one | Generator | | 6b-Hydroxytestosterone | Generator | | 6β-hydroxytestosterone | Generator | | 4-Androsten-6b,17b-diol-3-one | Generator | | 4-Androsten-6β,17β-diol-3-one | Generator | | 6b,17b-Dihydroxy-4-androsten-3-one | Generator | | 6β,17β-dihydroxy-4-androsten-3-one | Generator | | 6b,17b-Dihydroxyandrost-4-en-3-one | Generator | | 6β,17β-dihydroxyandrost-4-en-3-one | Generator | | 6 beta Hydroxy testosterone | HMDB | | 6,17-Dihydroxy-(6b,17b)-androst-4-en-3-one | HMDB | | 6,17-Dihydroxy-(6beta,17beta)-androst-4-en-3-one | HMDB | | 6,17-Dihydroxyandrost-4-en-3-one (acd/name 4.0) | HMDB | | 6 beta-Hydroxytestosterone, (17beta)-isomer | MeSH, HMDB | | 6 beta-Hydroxytestosterone | MeSH, HMDB | | 6 beta-Hydroxytestosterone, (6alpha,17beta)-isomer | MeSH, HMDB |
|
|---|
| Chemical Formula | C19H28O3 |
|---|
| Average Molecular Mass | 304.424 g/mol |
|---|
| Monoisotopic Mass | 304.204 g/mol |
|---|
| CAS Registry Number | 62-99-7 |
|---|
| IUPAC Name | (1S,2R,8R,10R,11S,14S,15S)-8,14-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadec-6-en-5-one |
|---|
| Traditional Name | 6β-hydroxytestosterone |
|---|
| SMILES | [H][C@@]12CC[C@H](O)[C@@]1(C)CC[C@@]1([H])[C@@]2([H])C[C@@H](O)C2=CC(=O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C19H28O3/c1-18-7-5-11(20)9-15(18)16(21)10-12-13-3-4-17(22)19(13,2)8-6-14(12)18/h9,12-14,16-17,21-22H,3-8,10H2,1-2H3/t12-,13-,14-,16+,17-,18+,19-/m0/s1 |
|---|
| InChI Key | XSEGWEUVSZRCBC-ZVBLRVHNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-alkyl,2-acylglycerols. These are glycerides consisting of two fatty acyl chains covalently bonded to a glycerol molecule at the 1- and 2-positions through an ether and an ester linkage, respectively. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Diradylglycerols |
|---|
| Direct Parent | 1-alkyl,2-acylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-alkyl,2-acylglycerol
- Fatty acid ester
- Glycerol ether
- Fatty acyl
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Ether
- Dialkyl ether
- Carboxylic acid derivative
- Organic oxide
- Primary alcohol
- Organooxygen compound
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002r-0490000000-fbce40562e85113343e0 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-001i-3201900000-19a880626afdcb624da4 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0092000000-489bed8773fcd0c13de6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-0090000000-f07fc698e115a5f1c7cb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pvl-3490000000-3c10f54e817114fb8e5a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0049000000-2030860ac7b87d34beb0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udr-0097000000-0f7ab0e063794ee5cfd2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dr-1190000000-d45875a741e3ea1e7526 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0019000000-9b79f62ea088c74b3a3d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-1794000000-57b269ff6c81d8c47be7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-3910000000-eed8482fba4fd9079a6f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-93986d59045cc7fce551 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0009000000-93986d59045cc7fce551 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gbi-0092000000-1bd122937767d0574fc2 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0011146 |
|---|
| FooDB ID | FDB027921 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 18558308 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 18603089 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|