| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:40:02 UTC |
|---|
| Update Date | 2016-11-09 01:17:32 UTC |
|---|
| Accession Number | CHEM022575 |
|---|
| Identification |
|---|
| Common Name | Aldophosphamide |
|---|
| Class | Small Molecule |
|---|
| Description | Detoxification of cyclophosphamide is effected, in part, by hepatic class 1 aldehyde dehydrogenase (ALDH-1)-catalyzed oxidation of aldophosphamide, a pivotal aldehyde intermediate, to the nontoxic metabolite, carboxyphosphamide. Detoxification of aldophosphamide may also be effected by enzymes, viz. Thus, NAD-linked oxidation and NADPH-linked reduction of aldophosphamide catalyzed by relevant erythrocyte enzymes were quantified. (PMID: 9394035) Class 1 aldehyde dehydrogenases (ALDH-1) function as drug resistance gene products by catalyzing the irreversible conversion of aldophosphamide, an active metabolite of cyclophosphamide, to an inert compound. (PMID: 9322086) |
|---|
| Contaminant Sources | - HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Cyclophosphamide-mustard | HMDB | | Cyp-mustard | HMDB |
|
|---|
| Chemical Formula | C7H15Cl2N2O3P |
|---|
| Average Molecular Mass | 277.085 g/mol |
|---|
| Monoisotopic Mass | 276.020 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
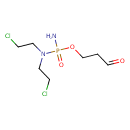
| IUPAC Name | 3-({amino[bis(2-chloroethyl)amino]phosphoryl}oxy)propanal |
|---|
| Traditional Name | aldophosphamide |
|---|
| SMILES | NP(=O)(OCCC=O)N(CCCl)CCCl |
|---|
| InChI Identifier | InChI=1S/C7H15Cl2N2O3P/c8-2-4-11(5-3-9)15(10,13)14-7-1-6-12/h6H,1-5,7H2,(H2,10,13) |
|---|
| InChI Key | QMGUSPDJTPDFSF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as nitrogen mustard compounds. Nitrogen mustard compounds are compounds having two beta-haloalkyl groups bound to a nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Nitrogen mustard compounds |
|---|
| Direct Parent | Nitrogen mustard compounds |
|---|
| Alternative Parents | |
|---|
| Substituents | - Nitrogen mustard
- Phosphoric monoester diamide
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Organic phosphoric acid amide
- Alpha-hydrogen aldehyde
- Organic oxide
- Organooxygen compound
- Organochloride
- Organopnictogen compound
- Organohalogen compound
- Organic oxygen compound
- Carbonyl group
- Aldehyde
- Alkyl halide
- Alkyl chloride
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01r6-3930000000-ff6f36c8b42295802040 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6r-3190000000-ab09c0d99d6be0494721 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9100000000-4f52a98720e48a89bf67 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvl-9300000000-b456c4d19d6e29b11aa4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1390000000-f2cfbeebb17c18444fc4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xr-9330000000-e296d1c2deb7425c67d6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udu-2910000000-ab68ba84f4c31e2560ee | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0390000000-85648cd89df23d147468 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0790000000-9db4ba9eb9886927554f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0btc-9100000000-d3145b5a45da48514901 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-6cd6113372d1d0808e1d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-2390000000-09ec42c4941802f6cb3d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pyi-4910000000-eb2eb8022150d34149a4 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0060433 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Cyclophosphamide |
|---|
| Chemspider ID | Not Available |
|---|
| ChEBI ID | 2560 |
|---|
| PubChem Compound ID | 107744 |
|---|
| Kegg Compound ID | C07645 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | ECMDB20115 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|