| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:36:28 UTC |
|---|
| Update Date | 2016-11-09 01:17:31 UTC |
|---|
| Accession Number | CHEM022476 |
|---|
| Identification |
|---|
| Common Name | Pyrazin-2-carboxylic acid |
|---|
| Class | Small Molecule |
|---|
| Description | The parent compound of the class of pyrazinecarboxylic acids, that is pyrazine bearing a single carboxy substituent. The active metabolite of the antitubercular drug pyrazinamide. |
|---|
| Contaminant Sources | - HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
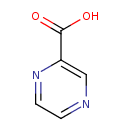
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Carboxypyrazine | ChEBI | | 2-Pyrazinecarboxylic acid | ChEBI | | Paradiazinecarboxylic acid | ChEBI | | Pyrazinecarboxylic acid | ChEBI | | Pyrazinemonocarboxylic acid | ChEBI | | Pyrazinic acid | ChEBI | | Pyrazinoic acid | ChEBI | | 2-Pyrazinecarboxylate | Generator | | Paradiazinecarboxylate | Generator | | Pyrazinecarboxylate | Generator | | Pyrazinemonocarboxylate | Generator | | Pyrazinate | Generator | | Pyrazinoate | Generator | | Pyrazin-2-carboxylate | Generator | | Pyrazine-2-carboxylic acid | MeSH | | Pyrazine-2-carboxylate | Generator |
|
|---|
| Chemical Formula | C5H4N2O2 |
|---|
| Average Molecular Mass | 124.098 g/mol |
|---|
| Monoisotopic Mass | 124.027 g/mol |
|---|
| CAS Registry Number | Not Available |
|---|
| IUPAC Name | pyrazine-2-carboxylic acid |
|---|
| Traditional Name | pyrazinoic acid |
|---|
| SMILES | OC(=O)C1=CN=CC=N1 |
|---|
| InChI Identifier | InChI=1S/C5H4N2O2/c8-5(9)4-3-6-1-2-7-4/h1-3H,(H,8,9) |
|---|
| InChI Key | NIPZZXUFJPQHNH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrazine carboxylic acids. These are heterocyclic compounds containing a pyrazine ring substituted by one or more carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyrazines |
|---|
| Direct Parent | Pyrazine carboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrazine carboxylic acid
- Heteroaromatic compound
- Azacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fc0-9100000000-0d1d9343efa8efa99226 | Spectrum | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fc0-9100000000-0d1d9343efa8efa99226 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ul0-9300000000-5d851f8b3e99335027a5 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fr-9700000000-dc1a24b248b553898896 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-9e64f2293e46344d81ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-2900000000-af86c6152381383e33e7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uk9-9000000000-cafc4cffd9f796c532e5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2900000000-576cc0731c2413e5e0d6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00fr-9800000000-d888658b0a8673ea88fe | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fvi-9000000000-eb742c3177bfe29f72e5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9000000000-90831ac2a79e645518ba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-90831ac2a79e645518ba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ufr-9000000000-11eb44f98f6f9300db58 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-2900000000-fc721474e0fad99dcba0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-9100000000-3bc909318a437df6cd91 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-c78d399db63d85709c22 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0059734 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | PYRAZINOIC-ACID |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Pyrazinoic_acid |
|---|
| Chemspider ID | 1018 |
|---|
| ChEBI ID | 71311 |
|---|
| PubChem Compound ID | 1047 |
|---|
| Kegg Compound ID | C19915 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | ECMDB02069 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=19298694 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=19963078 | | 3. https://www.ncbi.nlm.nih.gov/pubmed/?term=20099263 | | 4. https://www.ncbi.nlm.nih.gov/pubmed/?term=20382083 | | 5. https://www.ncbi.nlm.nih.gov/pubmed/?term=20429806 | | 6. https://www.ncbi.nlm.nih.gov/pubmed/?term=20879713 | | 7. https://www.ncbi.nlm.nih.gov/pubmed/?term=21283666 | | 8. https://www.ncbi.nlm.nih.gov/pubmed/?term=21775138 | | 9. https://www.ncbi.nlm.nih.gov/pubmed/?term=21835980 | | 10. https://www.ncbi.nlm.nih.gov/pubmed/?term=21876062 | | 11. https://www.ncbi.nlm.nih.gov/pubmed/?term=22004792 | | 12. https://www.ncbi.nlm.nih.gov/pubmed/?term=22074423 | | 13. https://www.ncbi.nlm.nih.gov/pubmed/?term=22236853 | | 14. https://www.ncbi.nlm.nih.gov/pubmed/?term=22372927 | | 15. https://www.ncbi.nlm.nih.gov/pubmed/?term=22613684 | | 16. https://www.ncbi.nlm.nih.gov/pubmed/?term=23101013 | | 17. https://www.ncbi.nlm.nih.gov/pubmed/?term=28166217 | | 18. Dolezal M, Zitko J, Kesetovicova D, Kunes J, Svobodova M: Substituted N-Phenylpyrazine-2-carboxamides: synthesis and antimycobacterial evaluation. Molecules. 2009 Oct 20;14(10):4180-9. doi: 10.3390/molecules14104180. | | 19. Aoki Y, Yoshida M, Kawaide H, Abe H, Natsume M: Structural determination of hypnosin, a spore germination inhibitor of phytopathogenic Streptomyces sp. causing root tumor in melon (Cucumis sp.). J Agric Food Chem. 2007 Dec 26;55(26):10622-7. Epub 2007 Dec 5. | | 20. Nakanishi T, Ohya K, Shimada S, Anzai N, Tamai I: Functional cooperation of URAT1 (SLC22A12) and URATv1 (SLC2A9) in renal reabsorption of urate. Nephrol Dial Transplant. 2013 Mar;28(3):603-11. doi: 10.1093/ndt/gfs574. Epub 2013 Jan 4. | | 21. Bansal G, Singh M, Jindal KC, Singh S: LC and LC-MS study on establishment of degradation pathway of glipizide under forced decomposition conditions. J Chromatogr Sci. 2008 Jul;46(6):510-7. | | 22. Vishweshwar P, Nangia A, Lynch VM: Recurrence of carboxylic acid-pyridine supramolecular synthon in the crystal structures of some pyrazinecarboxylic acids. J Org Chem. 2002 Jan 25;67(2):556-65. | | 23. Ma C, Han Y, Zhang R, Wang D: Self-assembly of diorganotin(IV) moieties and 2-pyrazinecarboxylic acid: syntheses, characterizations and crystal structures of monomeric, polymeric or trinuclear macrocyclic compounds. Dalton Trans. 2004 Jun 21;(12):1832-40. Epub 2004 May 13. | | 24. Dolezal M, Kesetovic D, Zitko J: Antimycobacterial evaluation of pyrazinoic acid reversible derivatives. Curr Pharm Des. 2011;17(32):3506-14. | | 25. Sato M, Wakayama T, Mamada H, Shirasaka Y, Nakanishi T, Tamai I: Identification and functional characterization of uric acid transporter Urat1 (Slc22a12) in rats. Biochim Biophys Acta. 2011 Jun;1808(6):1441-7. doi: 10.1016/j.bbamem.2010.11.002. Epub 2010 Nov 11. | | 26. Holzer W, Eller GA, Datterl B, Habicht D: Derivatives of pyrazinecarboxylic acid: 1H, 13C and 15N NMR spectroscopic investigations. Magn Reson Chem. 2009 Jul;47(7):617-24. doi: 10.1002/mrc.2437. | | 27. Nayak S, Harms K, Dehnen S: New three-dimensional metal-organic framework with heterometallic [Fe-Ag] building units: synthesis, crystal structure, and functional studies. Inorg Chem. 2011 Apr 4;50(7):2714-6. doi: 10.1021/ic1019636. Epub 2011 Mar 1. | | 28. Tanase S, Marques Gallego P, Bouwman E, Long GJ, Rebbouh L, Grandjean F, de Gelder R, Mutikainen I, Turpeinen U, Reedijk J: Versatility in the binding of 2-pyrazinecarboxylate with iron. Synthesis, structure and magnetic properties of iron(II) and iron(III) complexes. Dalton Trans. 2006 Apr 7;(13):1675-84. Epub 2005 Dec 15. | | 29. Dolezal M, Palek L, Vinsova J, Buchta V, Jampilek J, Kralova K: Substituted pyrazinecarboxamides: synthesis and biological evaluation. Molecules. 2006 Mar 29;11(4):242-56. | | 30. Sato M, Mamada H, Anzai N, Shirasaka Y, Nakanishi T, Tamai I: Renal secretion of uric acid by organic anion transporter 2 (OAT2/SLC22A7) in human. Biol Pharm Bull. 2010;33(3):498-503. | | 31. Servusova B, Eibinova D, Dolezal M, Kubicek V, Paterova P, Pesko M, Kralova K: Substituted N-benzylpyrazine-2-carboxamides: synthesis and biological evaluation. Molecules. 2012 Nov 6;17(11):13183-98. doi: 10.3390/molecules171113183. | | 32. Cuevas A, Kremer C, Hummert M, Schumann H, Lloret F, Julve M, Faus J: Magnetic properties and molecular structures of binuclear (2-pyrazinecarboxylate)-bridged complexes containing Re(IV) and M(II) (M = Co, Ni). Dalton Trans. 2007 Jan 21;(3):342-50. Epub 2006 Nov 21. | | 33. Dolezal M, Zitko J, Osicka Z, Kunes J, Vejsova M, Buchta V, Dohnal J, Jampilek J, Kralova K: Synthesis, antimycobacterial, antifungal and photosynthesis-inhibiting activity of chlorinated N-phenylpyrazine-2-carboxamides. Molecules. 2010 Nov 26;15(12):8567-81. doi: 10.3390/molecules15128567. | | 34. Knope KE, Kimura H, Yasaka Y, Nakahara M, Andrews MB, Cahill CL: Investigation of in situ oxalate formation from 2,3-pyrazinedicarboxylate under hydrothermal conditions using nuclear magnetic resonance spectroscopy. Inorg Chem. 2012 Mar 19;51(6):3883-90. doi: 10.1021/ic3000944. Epub 2012 Feb 24. |
|
|---|