| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:35:15 UTC |
|---|
| Update Date | 2016-11-09 01:17:30 UTC |
|---|
| Accession Number | CHEM022447 |
|---|
| Identification |
|---|
| Common Name | 3,7-Dimethyluric acid |
|---|
| Class | Small Molecule |
|---|
| Description | An oxopurine that is 7,9-dihydro-1H-purine-2,6,8(3H)-trione substituted by methyl groups at N-3 and N-7. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
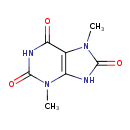
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,7-Dimethylate | Generator | | 3,7-Dimethylic acid | Generator | | 37-Dimethyluric acid | ChEMBL, HMDB | | 37-Dimethylate | Generator, HMDB | | 37-Dimethylic acid | Generator, HMDB | | 3,7-Dimethyl-2,6,8-trihydroxypurine | HMDB | | 3,7-Dimethyl-7,9-dihydro-1H-purine-2,6,8(3H)-trione | HMDB | | 3,7-DMU | MeSH, HMDB |
|
|---|
| Chemical Formula | C7H8N4O3 |
|---|
| Average Molecular Mass | 196.163 g/mol |
|---|
| Monoisotopic Mass | 196.060 g/mol |
|---|
| CAS Registry Number | 13087-49-5 |
|---|
| IUPAC Name | 3,7-dimethyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 3,7-dimethyluric acid |
|---|
| SMILES | CN1C(=O)NC2=C1C(=O)NC(=O)N2C |
|---|
| InChI Identifier | InChI=1S/C7H8N4O3/c1-10-3-4(8-6(10)13)11(2)7(14)9-5(3)12/h1-2H3,(H,8,13)(H,9,12,14) |
|---|
| InChI Key | HMLZLHKHNBLLJD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0v0a-0900000000-46adc1bd1a2829ed3eab | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-0900000000-5554059238ba6f5a4aa7 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001l-9800000000-bcef3be559d5d3540d62 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-014l-9000000000-7d225864dafe7484ef17 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-81b25f479c4f1663781d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0007-0900000000-0f875011e8751037af3f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-ddcf3e38bdc4d7198abc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-f9c0039adcc98c79f10d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f6t-0900000000-5398ccddcd1ad6de4811 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-79381deb2ab0e04daf89 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-0e499ff6f23e328dc54a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0900000000-c0e1ff53101ddbba850d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9300000000-a0758f9ce9eb83d7a97d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-8ba3918ee48755b0ba1a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-2900000000-a5948ea7307823697f8e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-82b93be35cae293c6cab | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0001982 |
|---|
| FooDB ID | FDB022780 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | CPD-12482 |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 74994 |
|---|
| ChEBI ID | 68531 |
|---|
| PubChem Compound ID | 83126 |
|---|
| Kegg Compound ID | C16360 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | M2MDB005818 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|