| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:30:49 UTC |
|---|
| Update Date | 2016-11-09 01:17:29 UTC |
|---|
| Accession Number | CHEM022387 |
|---|
| Identification |
|---|
| Common Name | Tiglylcarnitine |
|---|
| Class | Small Molecule |
|---|
| Description | An O-acyl-L-carnitine compound having trans-2-methyl-2-butenoyl (tiglyl) as the acyl substituent. |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
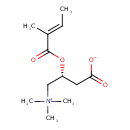
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-Tiglyl-carnitine | ChEBI | | O-Tiglylcarnitine | HMDB | | O-Tiglyl-L-carnitine | HMDB | | Tiglyl carnitine | HMDB | | Tiglylcarnitine | HMDB |
|
|---|
| Chemical Formula | C12H21NO4 |
|---|
| Average Molecular Mass | 243.299 g/mol |
|---|
| Monoisotopic Mass | 243.147 g/mol |
|---|
| CAS Registry Number | 64191-86-2 |
|---|
| IUPAC Name | (3R)-3-{[(2E)-2-methylbut-2-enoyl]oxy}-4-(trimethylazaniumyl)butanoate |
|---|
| Traditional Name | (3R)-3-{[(2E)-2-methylbut-2-enoyl]oxy}-4-(trimethylammonio)butanoate |
|---|
| SMILES | C\C=C(/C)C(=O)OC(CC([O-])=O)C[N+](C)(C)C |
|---|
| InChI Identifier | InChI=1S/C12H21NO4/c1-6-9(2)12(16)17-10(7-11(14)15)8-13(3,4)5/h6,10H,7-8H2,1-5H3/b9-6+ |
|---|
| InChI Key | WURBQCVBQNMUQT-RMKNXTFCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyl carnitines. These are organic compounds containing a fatty acid with the carboxylic acid attached to carnitine through an ester bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acid esters |
|---|
| Direct Parent | Acyl carnitines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acyl-carnitine
- Branched fatty acid
- Dicarboxylic acid or derivatives
- Tetraalkylammonium salt
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- Quaternary ammonium salt
- Carboxylic acid ester
- Carboxylic acid salt
- Carboxylic acid derivative
- Carboxylic acid
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Organic salt
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0090000000-e78bd389593472138484 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000l-9050000000-1bb9d023b199264c5fe7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-e9262cbaff8cb4ad0ba6 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0002366 |
|---|
| FooDB ID | FDB022980 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 34999729 |
|---|
| ChEBI ID | 85520 |
|---|
| PubChem Compound ID | 91825636 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | YMDB01537 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Fontaine M, Briand G, Ser N, Armelin I, Rolland MO, Degand P, Vamecq J: Metabolic studies in twin brothers with 2-methylacetoacetyl-CoA thiolase deficiency. Clin Chim Acta. 1996 Nov 15;255(1):67-83. | | 2. Fukao T, Zhang GX, Sakura N, Kubo T, Yamaga H, Hazama A, Kohno Y, Matsuo N, Kondo M, Yamaguchi S, Shigematsu Y, Kondo N: The mitochondrial acetoacetyl-CoA thiolase (T2) deficiency in Japanese patients: urinary organic acid and blood acylcarnitine profiles under stable conditions have subtle abnormalities in T2-deficient patients with some residual T2 activity. J Inherit Metab Dis. 2003;26(5):423-31. | | 3. Millington DS, Roe CR, Maltby DA: Characterization of new diagnostic acylcarnitines in patients with beta-ketothiolase deficiency and glutaric aciduria type I using mass spectrometry. Biomed Environ Mass Spectrom. 1987 Dec;14(12):711-6. | | 4. Violante S, Ijlst L, Ruiter J, Koster J, van Lenthe H, Duran M, de Almeida IT, Wanders RJ, Houten SM, Ventura FV: Substrate specificity of human carnitine acetyltransferase: Implications for fatty acid and branched-chain amino acid metabolism. Biochim Biophys Acta. 2013 Jun;1832(6):773-9. doi: 10.1016/j.bbadis.2013.02.012. Epub 2013 Feb 24. | | 5. Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. | | 6. FRITZ IB: Action of carnitine on long chain fatty acid oxidation by liver. Am J Physiol. 1959 Aug;197:297-304. doi: 10.1152/ajplegacy.1959.197.2.297. | | 7. Makarova E, Makrecka-Kuka M, Vilks K, Volska K, Sevostjanovs E, Grinberga S, Zarkova-Malkova O, Dambrova M, Liepinsh E: Decreases in Circulating Concentrations of Long-Chain Acylcarnitines and Free Fatty Acids During the Glucose Tolerance Test Represent Tissue-Specific Insulin Sensitivity. Front Endocrinol (Lausanne). 2019 Dec 17;10:870. doi: 10.3389/fendo.2019.00870. eCollection 2019. | | 8. Kiykim E, Aktuglu Zeybek AC, Barut K, Zubarioglu T, Cansever MS, Alsancak S, Kasapcopur O: Screening of Free Carnitine and Acylcarnitine Status in Children With Familial Mediterranean Fever. Arch Rheumatol. 2016 Mar 10;31(2):133-138. doi: 10.5606/ArchRheumatol.2016.5696. eCollection 2016 Jun. | | 9. Wen P, Chen Z, Wang G, Su Z, Zhang X, Tang G, Cui D, Liu X, Li C: [Analysis of clinical phenotype and ACAT1 gene mutation in a family affected with beta-ketothiolase deficiency]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2016 Jun;33(3):286-91. doi: 10.3760/cma.j.issn.1003-9406.2016.03.002. | | 10. Pajares S, Lopez RM, Gort L, Argudo-Ramirez A, Marin JL, Gonzalez de Aledo-Castillo JM, Garcia-Villoria J, Arranz JA, Del Toro M, Tort F, Ugarteburu O, Casellas MD, Fernandez R, Ribes A: An incidental finding in newborn screening leading to the diagnosis of a patient with ECHS1 mutations. Mol Genet Metab Rep. 2020 Jan 2;22:100553. doi: 10.1016/j.ymgmr.2019.100553. eCollection 2020 Mar. | | 11. Wu H, Chen Y, Li Z, Liu X: Untargeted metabolomics profiles delineate metabolic alterations in mouse plasma during lung carcinoma development using UPLC-QTOF/MS in MS(E) mode. R Soc Open Sci. 2018 Sep 19;5(9):181143. doi: 10.1098/rsos.181143. eCollection 2018 Sep. | | 12. Yu ZR, Ning Y, Yu H, Tang NJ: A HPLC-Q-TOF-MS-based urinary metabolomic approach to identification of potential biomarkers of metabolic syndrome. J Huazhong Univ Sci Technolog Med Sci. 2014 Apr;34(2):276-283. doi: 10.1007/s11596-014-1271-7. Epub 2014 Apr 8. | | 13. Dambrova M, Makrecka-Kuka M, Kuka J, Vilskersts R, Nordberg D, Attwood MM, Smesny S, Sen ZD, Guo AC, Oler E, Tian S, Zheng J, Wishart DS, Liepinsh E, Schioth HB: Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol Rev. 2022 Jul;74(3):506-551. doi: 10.1124/pharmrev.121.000408. |
|
|---|