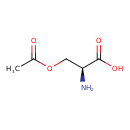
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-01c0-0900000000-cbac2ae436dcca0d0251 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-01c0-0900000000-7c5ef6cc55e1a01984af | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-00di-6900000000-f2863fb0291664c5ca21 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-00yi-0910000000-4eb5131c64279fbabbfb | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-01c0-0900000000-cbac2ae436dcca0d0251 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-01c0-0900000000-7c5ef6cc55e1a01984af | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-6900000000-f2863fb0291664c5ca21 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-00yi-0910000000-4eb5131c64279fbabbfb | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9000000000-1a24fb3faa274e72fcc5 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-006x-9300000000-048d27b5feab5a974f8d | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-052r-9700000000-798191f65808f670b0b8 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-052r-9700000000-798191f65808f670b0b8 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 35V, negative | splash10-0ai0-9000000000-48b5a588a0a890044838 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00di-9400000000-6ea6ba6b54ea90c56416 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-05gj-9400000000-32253d09352cdfb8c318 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-022l-9000000000-155f852f94fd442482f2 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-e88b0e868a77f604b59c | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0089-9100000000-c7a43c74f2dde018ff1b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00dj-9600000000-304c64ed85255dae12e8 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0btl-9400000000-7aa1b76f2e2dc6086311 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-01ox-9000000000-fd8ce6d36426fce8d343 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-01bc-9000000000-abc90d9cacd4b889cd96 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0002-2900000000-d04884f0db647a14fc8d | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-052r-8900000000-c8515198104000e95777 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0002-4900000000-1942c426e3cf9c8b41c6 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00di-9400000000-8d3a49712627498e2c9b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-052r-9700000000-cf8d37dfdbdee7d76628 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-052r-9700000000-32279928e0b151d0e1da | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0002-6900000000-cb1c33a71a73963cf0c1 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uea-3900000000-ef46289d46aa69d40d48 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udr-9700000000-221b830dc83fdf079527 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-52a79cff992a3a467f06 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-6900000000-4e0de865dfcc2446fe14 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9300000000-5a3f4b66b56fbaca505c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-123b2507eee8f6a9f064 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |