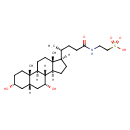
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053r-0212900000-197acffecf402e598644 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-004i-4214329000-11c176071503161e2cd4 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_6) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-02ar-0006900000-53366e607f8bd2e50783 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0og0-1595100000-2488fbc0ad2dab225684 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0000920000-d308c77003ce029da322 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000900000-4f77a6fb2b33e4e1a35f | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-671f53e46ce019da43dc | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0og0-1595100000-6670e8e89de5e9a0c3a1 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0000920000-0b63233ca30c95bee595 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0000900000-54b7fff092e1f9db6a20 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0002-0000900000-f978cbaedb898eee1843 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00e9-0801920000-ecfc97935416325b3ca5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-2901100000-9a3cf7f2963c041be363 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-8904100000-98cfa2606dbba4ef47a0 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000t-2001900000-4adf0e40de3812e8d0ba | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-6404900000-4d05b6f0352aebf4228d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-9102000000-63892e6e06f1123f8bfc | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-0000970000-8561365963466c5db093 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bvi-3509710000-451e758e0b96f9fa8ee7 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00l5-9463000000-16d35ecb5101cbd28ea9 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-31f17d1d48b7afdf3bae | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0000900000-a426f175803c68cb8313 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000t-7400900000-5a469259d1091a858347 | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |