| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:13:24 UTC |
|---|
| Update Date | 2016-11-09 01:17:22 UTC |
|---|
| Accession Number | CHEM021823 |
|---|
| Identification |
|---|
| Common Name | Ursocholic acid |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
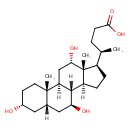
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3alpha,5beta,7beta,12alpha)-3,7,12-Trihydroxycholan-24-Oic acid | ChEBI | | 3alpha,7beta,12alpha-Trihydroxy-5beta-cholan-24-Oic acid | ChEBI | | 3alpha,7beta,12alpha-Trihydroxy-5beta-cholanic acid | ChEBI | | 7-Epicholic acid | ChEBI | | 7beta-Hydroxyisocholic acid | Kegg | | (3a,5b,7b,12a)-3,7,12-Trihydroxycholan-24-Oate | Generator | | (3a,5b,7b,12a)-3,7,12-Trihydroxycholan-24-Oic acid | Generator | | (3alpha,5beta,7beta,12alpha)-3,7,12-Trihydroxycholan-24-Oate | Generator | | (3Α,5β,7β,12α)-3,7,12-trihydroxycholan-24-Oate | Generator | | (3Α,5β,7β,12α)-3,7,12-trihydroxycholan-24-Oic acid | Generator | | 3a,7b,12a-Trihydroxy-5b-cholan-24-Oate | Generator | | 3a,7b,12a-Trihydroxy-5b-cholan-24-Oic acid | Generator | | 3alpha,7beta,12alpha-Trihydroxy-5beta-cholan-24-Oate | Generator | | 3Α,7β,12α-trihydroxy-5β-cholan-24-Oate | Generator | | 3Α,7β,12α-trihydroxy-5β-cholan-24-Oic acid | Generator | | 3a,7b,12a-Trihydroxy-5b-cholanate | Generator | | 3a,7b,12a-Trihydroxy-5b-cholanic acid | Generator | | 3alpha,7beta,12alpha-Trihydroxy-5beta-cholanate | Generator | | 3Α,7β,12α-trihydroxy-5β-cholanate | Generator | | 3Α,7β,12α-trihydroxy-5β-cholanic acid | Generator | | 7-Epicholate | Generator | | 7b-Hydroxyisocholate | Generator | | 7b-Hydroxyisocholic acid | Generator | | 7beta-Hydroxyisocholate | Generator | | 7Β-hydroxyisocholate | Generator | | 7Β-hydroxyisocholic acid | Generator | | Ursocholate | Generator | | 3a,7b,12a-Trihydroxy-5b-cholanoate | HMDB | | 3a,7b,12a-Trihydroxy-5b-cholanoic acid | HMDB | | 3a,7b,12a-Trihydroxycholanate | HMDB | | 3a,7b,12a-Trihydroxycholanic acid | HMDB | | 3 alpha,7 beta,12 alpha-Trihydroxy-5 beta-cholan-24-Oic acid | HMDB |
|
|---|
| Chemical Formula | C24H40O5 |
|---|
| Average Molecular Mass | 408.571 g/mol |
|---|
| Monoisotopic Mass | 408.288 g/mol |
|---|
| CAS Registry Number | 2955-27-3 |
|---|
| IUPAC Name | (4R)-4-[(1S,2S,5R,7S,9S,10R,11S,14R,15R,16S)-5,9,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | ursocholic acid |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)CCC(O)=O)[C@@]1(C)[C@@H](O)C[C@@]1([H])[C@@]2([H])[C@@H](O)C[C@]2([H])C[C@H](O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C24H40O5/c1-13(4-7-21(28)29)16-5-6-17-22-18(12-20(27)24(16,17)3)23(2)9-8-15(25)10-14(23)11-19(22)26/h13-20,22,25-27H,4-12H2,1-3H3,(H,28,29)/t13-,14+,15-,16-,17+,18+,19+,20+,22+,23+,24-/m1/s1 |
|---|
| InChI Key | BHQCQFFYRZLCQQ-UTLSPDKDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Trihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 12-hydroxysteroid
- 7-hydroxysteroid
- 7-alpha-hydroxysteroid
- 3-alpha-hydroxysteroid
- Hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Polyol
- Monocarboxylic acid or derivatives
- Organic oxide
- Alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03ec-0439000000-abd2a5c162e23ea0d78f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-001i-1100049000-ae391322cbbc458fdb46 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-1019800000-4a5ffc11cb3fa93e46cb | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0000900000-7f097ee7b44073c18648 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-07966770f57ff86aa177 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0009100000-915584a506714c470270 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0009000000-f970a088fc3d9cdb8fe6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-017j-3109000000-bb054743c390ef266044 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0008900000-ed202aebd950e99a7852 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-1009300000-db2a4b9107b891a8ef3a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9006000000-87df039839010127a1a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0001900000-450d1e4d25aae2ed99f4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-0005900000-630ea179fce67da6f208 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0r09-0019200000-0754019de78fe2be314d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0abc-0009400000-fe4f5c75dd7ef6e8d9ce | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-059l-3159100000-f0eff49c7e8da5d00c58 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-9640000000-28284141ad8f72ab8fe1 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0000917 |
|---|
| FooDB ID | FDB022316 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | 5871 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 109088 |
|---|
| ChEBI ID | 81240 |
|---|
| PubChem Compound ID | 122340 |
|---|
| Kegg Compound ID | C17644 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Terada, Masaki; Sato, Manabu; Horie, Yoshiaki; Takatsu, Mitsumune; Minami, Junichi. Production of ursocholic acid. Jpn. Kokai Tokkyo Koho (1986), 9 pp. | | 2. Batta AK, Salen G, Abroon J: Ursocholic acid, a hydrophilic bile acid, fails to improve liver function parameters in primary biliary cirrhosis: comparison with ursodeoxycholic acid. Am J Gastroenterol. 1997 Jun;92(6):1035-7. | | 3. Lanzini A, Pigozzi G, Facchinetti D, Bettini L, Castellano M, Beschi M, Rossi A, Muiesan G: Effect of chronic ursocholic acid administration on bile lipid composition and bile acid pool size in gallstone patients. Scand J Gastroenterol. 1990 Jul;25(7):711-9. | | 4. Tint GS, Batta AK, Dayal B, Kovell N, Shefer S, Salen G: Metabolism of ursocholic acid in humans: conversion of ursocholic acid to deoxycholic acid. Hepatology. 1992 Apr;15(4):645-50. | | 5. Li H, Chen F, Shang Q, Pan L, Shneider BL, Chiang JY, Forman BM, Ananthanarayanan M, Tint GS, Salen G, Xu G: FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am J Physiol Gastrointest Liver Physiol. 2005 Jan;288(1):G60-6. | | 6. Loria P, Carulli N, Medici G, Menozzi D, Salvioli G, Bertolotti M, Montanari M: Effect of ursocholic acid on bile lipid secretion and composition. Gastroenterology. 1986 Apr;90(4):865-74. | | 7. Nakashima T, Sakamoto Y, Inaba K, Mitsuyoshi H, Ishikawa H, Nakajima Y, Sakai M, Shima T, Kashima K: A paucity of unusual trihydroxy bile acids in the urine of patients with severe liver diseases. Hepatology. 1999 May;29(5):1518-22. | | 8. St-Pierre MV, Kullak-Ublick GA, Hagenbuch B, Meier PJ: Transport of bile acids in hepatic and non-hepatic tissues. J Exp Biol. 2001 May;204(Pt 10):1673-86. | | 9. Claudel T, Staels B, Kuipers F: The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005 Oct;25(10):2020-30. Epub 2005 Jul 21. | | 10. Chiang JY: Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003 Mar;284(3):G349-56. | | 11. Davis RA, Miyake JH, Hui TY, Spann NJ: Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002 Apr;43(4):533-43. | | 12. https://www.ncbi.nlm.nih.gov/pubmed/?term=10216137 | | 13. https://www.ncbi.nlm.nih.gov/pubmed/?term=12401785 | | 14. https://www.ncbi.nlm.nih.gov/pubmed/?term=1551642 | | 15. https://www.ncbi.nlm.nih.gov/pubmed/?term=1943496 | | 16. https://www.ncbi.nlm.nih.gov/pubmed/?term=2079611 | | 17. https://www.ncbi.nlm.nih.gov/pubmed/?term=22198717 | | 18. https://www.ncbi.nlm.nih.gov/pubmed/?term=2396085 | | 19. https://www.ncbi.nlm.nih.gov/pubmed/?term=24076126 | | 20. https://www.ncbi.nlm.nih.gov/pubmed/?term=26718089 | | 21. https://www.ncbi.nlm.nih.gov/pubmed/?term=28751127 | | 22. https://www.ncbi.nlm.nih.gov/pubmed/?term=3660436 | | 23. https://www.ncbi.nlm.nih.gov/pubmed/?term=3715370 | | 24. https://www.ncbi.nlm.nih.gov/pubmed/?term=3949116 | | 25. https://www.ncbi.nlm.nih.gov/pubmed/?term=7676478 | | 26. https://www.ncbi.nlm.nih.gov/pubmed/?term=8500738 | | 27. https://www.ncbi.nlm.nih.gov/pubmed/?term=9177526 |
|
|---|