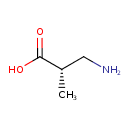
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9000000000-59e52601f90d8e636d61 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00ar-9200000000-0eb92539979a9fa36dbd | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-0udi-1900000000-aaccebe5d4a868bc32bf | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-0udi-2900000000-4f4e49fe169d5c21efc1 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-0udi-4900000000-f32e82420f5efb60456e | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-0uk9-6900000000-cb0ef82b397e24372142 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-0uk9-8900000000-31fc31847c8771693d15 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-0fk9-9600000000-6379643466e30660a05b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-00di-9300000000-97e8a7125780e41fceaf | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-05fr-9200000000-8f0488fe62603d121536 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-0ab9-9100000000-84e07fbee3e332502265 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 6V, negative | splash10-0a4i-9000000000-3634b730902e2e080b56 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - n/a 7V, negative | splash10-00di-9000000000-1b9078e3902a11473fa0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, positive | splash10-0udi-1900000000-76d19f44560e6b1b6f80 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, positive | splash10-0udi-1900000000-bd4e85f35b448a2b7540 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, positive | splash10-0udi-2900000000-b1371ec53be10ec9c411 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, positive | splash10-0udi-3900000000-d16b3475fcdfed5038d5 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, positive | splash10-0udi-4900000000-cd34930bbcb805c7525f | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, positive | splash10-0udr-5900000000-4b73ac1046680185a981 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, positive | splash10-0udr-6900000000-a911da27982a560a6cf1 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, positive | splash10-0udr-8900000000-91a255aae27e4e4aabb8 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-9300000000-30669ed04aadf3d3f040 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000l-9000000000-ca65637476e64152c44e | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-04d1648391d3f90db226 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-3900000000-ec54fcce079024a62344 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zfr-9500000000-0394f399cdf5df93c5df | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-d0c495c908ed905558a2 | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |