| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-22 07:08:42 UTC |
|---|
| Update Date | 2016-11-09 01:16:12 UTC |
|---|
| Accession Number | CHEM020482 |
|---|
| Identification |
|---|
| Common Name | O6-Methylguanine |
|---|
| Class | Small Molecule |
|---|
| Description | A methylguanine in which the methyl group is positioned on the oxygen at position 6. Formed in DNA by alkylation of the oxygen atom of guanine, most often by N-nitroso compounds and sometimes due to methylation by other compounds such as endogenous S-adenosylmethionine, it base-pairs to thymine rather than cytidine, causing a G:C to A:T transition in DNA. |
|---|
| Contaminant Sources | - ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
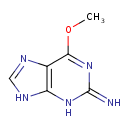
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-Methoxy-1H-purine-2-amine | ChEBI | | 6-Methoxyguanine | ChEBI | | O-(6)-Methylguanine | ChEBI | | O(6)-Methylguanine | ChEBI | | O6-Methylguanine | HMDB | | 6-Methylguanine | HMDB | | 2-amino-6-Methoxypurine | ChEBI |
|
|---|
| Chemical Formula | C6H7N5O |
|---|
| Average Molecular Mass | 165.153 g/mol |
|---|
| Monoisotopic Mass | 165.065 g/mol |
|---|
| CAS Registry Number | 20535-83-5 |
|---|
| IUPAC Name | 6-methoxy-3,9-dihydro-2H-purin-2-imine |
|---|
| Traditional Name | 6-O-methylguanine |
|---|
| SMILES | COC1=NC(=N)NC2=C1N=CN2 |
|---|
| InChI Identifier | InChI=1S/C6H7N5O/c1-12-5-3-4(9-2-8-3)10-6(7)11-5/h2H,1H3,(H3,7,8,9,10,11) |
|---|
| InChI Key | BXJHWYVXLGLDMZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hypoxanthines. Hypoxanthines are compounds containing the purine derivative 1H-purin-6(9H)-one. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Hypoxanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hypoxanthine
- Alkyl aryl ether
- Aminopyrimidine
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Ether
- Azacycle
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organopnictogen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00m0-2900000000-398edee1515b94d58cdb | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-f5ef9704d5e397cd7449 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-dcd4b3406016ec545ecf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-2900000000-ecdc42de6a212c28bdb0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-44b8935cd0a14254518b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03e9-0900000000-2dc830f7eebc50ea3071 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6u-9100000000-fef04240513f89db1ceb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-7f20cf9333bbb1eb98e1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-20ce136b99632e9d59a8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9100000000-e0df9e6b94989688a4d0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-11e5406fb2e0acd600c0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03e9-1900000000-992be0859a60c1e24588 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066r-9300000000-8aa422a3ae4b12fd0736 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0245002 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | 6-O-Methylguanine |
|---|
| Chemspider ID | 58766 |
|---|
| ChEBI ID | 20689 |
|---|
| PubChem Compound ID | 65275 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|