| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-22 06:36:07 UTC |
|---|
| Update Date | 2016-11-09 01:16:05 UTC |
|---|
| Accession Number | CHEM019878 |
|---|
| Identification |
|---|
| Common Name | Paricalcitol |
|---|
| Class | Small Molecule |
|---|
| Description | Paricalcitol is a synthetic vitamin D analog. Paricalcitol has been used to reduce parathyroid hormone levels. Paricalcitol is indicated for the prevention and treatment of secondary hyperparathyroidism associated with chronic renal failure. |
|---|
| Contaminant Sources | - HMDB Contaminants - Urine
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
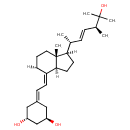
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 19-Nor-1alpha,25-dihydroxyvitamin D2 | ChEBI | | Zemplar | ChEBI | | 19-Nor-1a,25-dihydroxyvitamin D2 | Generator | | 19-Nor-1α,25-dihydroxyvitamin D2 | Generator | | Paricalcitol-D6 | HMDB | | 19-Nor-1,25-(OH)2D2 | HMDB | | Abbott brand OF paricalcitol | HMDB |
|
|---|
| Chemical Formula | C27H44O3 |
|---|
| Average Molecular Mass | 416.637 g/mol |
|---|
| Monoisotopic Mass | 416.329 g/mol |
|---|
| CAS Registry Number | 131918-61-1 |
|---|
| IUPAC Name | (1R,3R)-5-{2-[(1R,3aS,4E,7aR)-1-[(2R,3E,5S)-6-hydroxy-5,6-dimethylhept-3-en-2-yl]-7a-methyl-octahydro-1H-inden-4-ylidene]ethylidene}cyclohexane-1,3-diol |
|---|
| Traditional Name | paricalcitol |
|---|
| SMILES | [H]C1CC[C@]2(C)[C@]([H])(CC[C@@]2([H])\C1=C\C=C1C[C@@H](O)C[C@H](O)C1)[C@H](C)\C=C\[C@H](C)C(O)(C)C |
|---|
| InChI Identifier | InChI=1S/C27H44O3/c1-18(8-9-19(2)26(3,4)30)24-12-13-25-21(7-6-14-27(24,25)5)11-10-20-15-22(28)17-23(29)16-20/h8-11,18-19,22-25,28-30H,6-7,12-17H2,1-5H3/b9-8+,21-11+/t18-,19+,22-,23-,24-,25+,27-/m1/s1 |
|---|
| InChI Key | BPKAHTKRCLCHEA-UBFJEZKGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as vitamin d and derivatives. Vitamin D and derivatives are compounds containing a secosteroid backbone, usually secoergostane or secocholestane. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Vitamin D and derivatives |
|---|
| Direct Parent | Vitamin D and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triterpenoid
- Tertiary alcohol
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-4029200000-0e69ac54ae775b70b07f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0159-1600329000-8c4425fcfb09a3a8bc15 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00l2-0119200000-4cbc3d3f58c0e0397be4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-055e-0349000000-a309ddc9a87435e13c61 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-3594000000-41ea81c5e5ebb9f76445 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0004900000-266949ec80702b2d054f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014j-1009500000-bd2d96a201a200fbe62e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05n1-8109000000-27fa7680d7b0ac0c65f8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0aba-1397100000-3a5eca81bbca5912cfba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-1293000000-2a86d8a15c43da0836bc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-6940000000-549359487e3e0fb6c62a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-67ea14fb7a26b9b2d929 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0003900000-0988c8886cb5d91453ff | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00ko-3957600000-a8f253c962035e908892 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00910 |
|---|
| HMDB ID | HMDB0015046 |
|---|
| FooDB ID | Not Available |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Paricalcitol |
|---|
| Chemspider ID | 4444552 |
|---|
| ChEBI ID | 7931 |
|---|
| PubChem Compound ID | 5281104 |
|---|
| Kegg Compound ID | C08127 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|