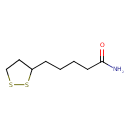
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-014i-6963000000-7e1887d53c2801b09961 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (1 TMS) | splash10-017l-3920000000-a3a7a08acf06d41c9b73 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-6963000000-7e1887d53c2801b09961 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-017l-3920000000-a3a7a08acf06d41c9b73 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-1910000000-b6ee336856e354af8114 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-3900000000-60cad9dd198c3ad64ed7 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f96-9800000000-ac967ad86beefc26c983 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-0900000000-24fca44fc025dbd6be18 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0ziu-9700000000-7dfd23f450bdfd581100 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-056u-9000000000-13583c4eb6b06a56c392 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-052r-2920000000-a4fb7176abf63e581f5e | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-000i-2900000000-10a079a84f024494ae23 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0zfr-9800000000-b01332176d6829b1eaa1 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0udl-9400000000-7a03c741edae36dff9cf | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0a5c-9100000000-7b44dfbfcbb0cc516254 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-000i-0900000000-50512172ce038bce2995 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-03di-0900000000-7652a33c17a37b6914aa | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0udi-0900000000-0df59a6540bede8a4a79 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-052r-2920000000-a4fb7176abf63e581f5e | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-000i-2900000000-158c946037c5d7bb3e50 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0zfr-9800000000-b01332176d6829b1eaa1 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0udl-9400000000-7a03c741edae36dff9cf | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a5c-9100000000-7b44dfbfcbb0cc516254 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-000i-0900000000-fa779b1c88469a732321 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-1900000000-9e4b6873b24c66cf9f69 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01p9-0900000000-b37ce6b25b9feef09b96 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4r-0940000000-d79e1aa52057e567d71f | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052r-3900000000-abd8148048c5f282d889 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0adr-9500000000-9769c619660ab6af46d2 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zml-1920000000-79d57d488cb8fbc804a3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fdo-4910000000-d264eaf6e242c36bc563 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-289d2f20046f7819a7ca | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |