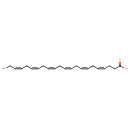
| Synonyms | | Value | Source |
|---|
| (4Z,7Z,10Z,13Z,16Z,19Z)-Docosahexaenoic acid | ChEBI | | 22:6(N-3) | ChEBI | | 22:6-4, 7,10,13,16,19 | ChEBI | | 4,7,10,13,16,19-Docosahexaenoic acid | ChEBI | | all-cis-4,7,10,13,16,19-Docosahexaenoic acid | ChEBI | | all-cis-DHA | ChEBI | | Cervonic acid | ChEBI | | DHA | ChEBI | | Doconexent | ChEBI | | DOCOSA-4,7,10,13,16,19-hexaenoIC ACID | ChEBI | | Docosahexaenoate | Kegg | | 4Z,7Z,10Z,13Z,16Z,19Z-Docosahexaenoic acid | Kegg | | (4Z,7Z,10Z,13Z,16Z,19Z)-Docosa-4,7,10,13,16,19-hexaenoic acid | Kegg | | (4Z,7Z,10Z,13Z,16Z,19Z)-Docosahexaenoate | Generator | | 4,7,10,13,16,19-Docosahexaenoate | Generator | | all-cis-4,7,10,13,16,19-Docosahexaenoate | Generator | | Cervonate | Generator | | DOCOSA-4,7,10,13,16,19-hexaenoate | Generator | | 4Z,7Z,10Z,13Z,16Z,19Z-Docosahexaenoate | Generator | | (4Z,7Z,10Z,13Z,16Z,19Z)-Docosa-4,7,10,13,16,19-hexaenoate | Generator | | all-Z-Docosahexaenoate | HMDB | | all-Z-Docosahexaenoic acid | HMDB | | cis-4,7,10,13,16,19-Docosahexanoate | HMDB | | cis-4,7,10,13,16,19-Docosahexanoic acid | HMDB | | Doconexento | HMDB | | Doconexentum | HMDB | | Doxonexent | HMDB | | Acids, docosahexaenoic | HMDB | | Acids, docosahexenoic | HMDB | | Docosahexaenoic acid, 4,7,10,13,16,19-(all-Z-isomer) | HMDB | | Docosahexaenoic acid (all-Z isomer) | HMDB | | Docosahexaenoic acid, 4,7,10,13,16,19-isomer | HMDB | | Efalex | HMDB | | (4Z,7Z,10Z,13Z,16Z,19Z)-4,7,10,13,16,19-Docosahexaenoic acid | HMDB | | (4Z,7Z,10Z,13Z,16Z,19Z)-Docosahexenoic acid | HMDB | | (all-Z)-4,7,10,13,16,19-Docosahexaenoic acid | HMDB | | 4-cis,7-cis,10-cis,13-cis,16-cis,19-cis-Docosahexaenoic acid | HMDB | | FA(22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | HMDB | | FA(22:6n3) | HMDB | | delta4,7,10,13,16,19-Docosahexaenoic acid | HMDB | | Δ4,7,10,13,16,19-docosahexaenoic acid | HMDB | | Docosahexaenoic acid | HMDB | | Choline docosahexaenoate | HMDB | | Choline docosahexaenoic acid | HMDB | | Docosahexaenoylcholine | HMDB |
|
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 TMS) | splash10-004l-9800000000-86f34228f9e92b6da2c6 | Spectrum | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-004l-9800000000-86f34228f9e92b6da2c6 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0690-5590000000-145821d84e425eec8f98 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0079-9363000000-db2668be2bbd1dbdd562 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-001i-0190000000-e6566b5aff7cefa4ae5d | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-0069000000-c76c91e0abd1895adb8b | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-001r-0069000000-a25e6a700612620a1217 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0ab9-9321000000-6b36fbd84ed730000eac | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-9642000000-5ecff42b9082b57b5be6 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-0389000000-737691a3fc2f16b3c59e | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-00l6-9510000000-8e81381f5f447b18e9fc | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0009000000-071caf7af6bdab525160 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1669000000-4e72cfd5cc68b32bd371 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0059-1179000000-289993a223b524e7c74f | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0019000000-aa9263466508c9f2e65c | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-004i-7389000000-4e6afa18acb803f01d23 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-9442000000-c452e1856fb3038c7f1e | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1569000000-4e72cfd5cc68b32bd371 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01t9-0139000000-7f2351e9d6b367bd3e6d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-3592000000-10365ee2a8e305e24bd4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ktf-8960000000-6382301474f6f5630982 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0019000000-f498ba345a6e0d89ae44 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-1069000000-be6f83d34620dceb650e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9130000000-abfb63ab3950fe4edf48 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03fr-1439000000-c5fd83b4d4f8531d17e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-3922000000-a59c9fe862e58a93dd03 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05o3-7900000000-c8cfc5620efa939690bb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0009000000-abcf527af18f1147a0aa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-1119000000-fbe7758a1c75272194e2 | Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-002f-9400000000-60bd7bfd4de9015476c7 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|