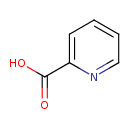
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-001r-0900000000-3d42fed18a7170ac902c | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (1 TMS) | splash10-005i-4900000000-ceebd42c8dcc23fdf3e4 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fb9-9000000000-bab2bcb7bd27b0be1f51 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-001r-0900000000-3d42fed18a7170ac902c | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-005i-4900000000-ceebd42c8dcc23fdf3e4 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-001r-0900000000-7276adde2578b546a8f2 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05i0-9400000000-7356d3453f3427cc9d89 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fr-9500000000-5d9f84aec4ab264b04db | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0adi-3900000000-7fac959b15471dc94406 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-004i-9000000000-0e912cf3a3db8f8ee5df | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0fb9-9000000000-6d29f5dc4ddc1bb1adb5 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - EI-B (MX-1303) , Positive | splash10-0fb9-9000000000-bab2bcb7bd27b0be1f51 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-05fr-0900000000-e169ce1e815eac2ad211 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-056s-9500000000-cd5fe946f692da4957ca | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-004i-9000000000-927c599edd5deaf854f0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-004i-9000000000-1d63eb2db32cd991a084 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-004i-9000000000-219a58c586353210f1a3 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-00di-0900000000-fe37c7a3e725d6a622fd | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-05fs-4900000000-b85a7afe0ba03853cfbf | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-05fr-0900000000-59ec49f98e7a39b33252 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-056s-9500000000-cd5fe946f692da4957ca | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9000000000-927c599edd5deaf854f0 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9000000000-1d63eb2db32cd991a084 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9000000000-219a58c586353210f1a3 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-00di-3900000000-9fd4a6ac15261e2c8d8e | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-02t9-9600000000-bc378bb03dd26ace9a39 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0900000000-fe37c7a3e725d6a622fd | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0900000000-b09ed1d66b009b4624ee | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-1900000000-a4d3409ab064e87942a9 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-ddbf2ed02e3b712aca5d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2900000000-7de5e81309a53ea848ef | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00fr-8900000000-0143384039159388b6eb | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-85d472523e404d49ec06 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |