| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-22 03:26:45 UTC |
|---|
| Update Date | 2016-10-28 10:02:23 UTC |
|---|
| Accession Number | CHEM016304 |
|---|
| Identification |
|---|
| Common Name | Ellagic acid |
|---|
| Class | Small Molecule |
|---|
| Description | An organic heterotetracyclic compound resulting from the formal dimerisation of gallic acid by oxidative aromatic coupling with intramolecular lactonisation of both carboxylic acid groups of the resulting biaryl. It is found in many fruits and vegetables, including raspberries, strawberries, cranberries, and pomegranates. |
|---|
| Contaminant Sources | - Cosmetic Chemicals
- FooDB Chemicals
- HMDB Contaminants - Urine
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
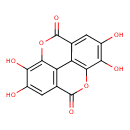
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,3,7,8-Tetrahydroxy[1]benzopyrano[5,4,3-cde][1]benzopyran-5,10-dione | ChEBI | | 4,4',5,5',6,6'-Hexahydroxydiphenic acid 2,6,2',6'-dilactone | ChEBI | | Acide ellagique | ChEBI | | Acido elagico | ChEBI | | Acidum ellagicum | ChEBI | | Benzoaric acid | ChEBI | | Ellagsaeure | ChEBI | | Lagistase | ChEBI | | 4,4',5,5',6,6'-Hexahydroxydiphenate 2,6,2',6'-dilactone | Generator | | Benzoarate | Generator | | Ellagate | Generator | | Acid, benzoaric | MeSH | | Acid, ellagic | MeSH | | Alizarine yellow | HMDB | | Elagostasine | HMDB | | Eleagate | HMDB | | Eleagic acid | HMDB | | Ellagic acid dihydrate | HMDB | | Ellagic acid hydrate | HMDB | | Gallogen | HMDB | | Llagate | HMDB | | Llagic acid | HMDB | | Ellagic acid | PhytoBank |
|
|---|
| Chemical Formula | C14H6O8 |
|---|
| Average Molecular Mass | 302.194 g/mol |
|---|
| Monoisotopic Mass | 302.006 g/mol |
|---|
| CAS Registry Number | 476-66-4 |
|---|
| IUPAC Name | 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),4,6,8(16),11,13-hexaene-3,10-dione |
|---|
| Traditional Name | 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),4,6,8(16),11,13-hexaene-3,10-dione |
|---|
| SMILES | OC1=C(O)C2=C3C(=C1)C(=O)OC1=C3C(=CC(O)=C1O)C(=O)O2 |
|---|
| InChI Identifier | InChI=1S/C14H6O8/c15-5-1-3-7-8-4(14(20)22-11(7)9(5)17)2-6(16)10(18)12(8)21-13(3)19/h1-2,15-18H |
|---|
| InChI Key | AFSDNFLWKVMVRB-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydrolyzable tannins. These are tannins with a structure characterized by either of the following models. In model 1, the structure contains galloyl units (in some cases, shikimic acid units) that are linked to diverse polyol carbohydrate-, catechin-, or triterpenoid units. In model 2, contains at least two galloyl units C-C coupled to each other, and do not contain a glycosidically linked catechin unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Phenylpropanoids and polyketides |
|---|
| Class | Tannins |
|---|
| Sub Class | Hydrolyzable tannins |
|---|
| Direct Parent | Hydrolyzable tannins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydrolyzable tannin
- Ellagic_acid
- 7,8-dihydroxycoumarin
- Coumarin
- Isocoumarin
- Benzopyran
- 2-benzopyran
- 1-benzopyran
- 1-hydroxy-2-unsubstituted benzenoid
- Pyranone
- Benzenoid
- Pyran
- Heteroaromatic compound
- Lactone
- Polyol
- Organoheterocyclic compound
- Oxacycle
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uk9-0095000000-fbb3052375510a43a32e | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0100-3000090000-98c2eaf6f099a674f556 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-0009000000-40fcade2aa63aa93a2ae | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0zi0-0192000000-778eac8f3ed58bcaac74 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - DI-ESI-qTof , Positive | splash10-0zi0-0093000000-8a6cc05fc65249815e82 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-0zfr-1589000000-a4f6e3a25012f7426f7d | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a6r-0192000000-39923f8b1dd96716ea79 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-001i-0090000000-a1f823c8fd710f0476d9 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0udi-0039000000-911cfbddb0cc618af274 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-0zfr-1589000000-a4f6e3a25012f7426f7d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0009000000-67c5a949d6d186ed40d5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0039000000-d2f61e8b5cdaa0f142ba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0090000000-961daa97dff240069d92 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0029000000-246b5797e3aacdc8f2f2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0059000000-cc29f1c85d471f50bb6b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-114i-0190000000-6c3c3d5d2c468609b612 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-9600b3c9d9dcbf3cca2a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0029000000-9f6b76087b40406d9ff3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0090000000-e7359efe114a8fa3c0e3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0009000000-49d4f264ad1b5c5b0fff | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0009000000-49d4f264ad1b5c5b0fff | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6s-0090000000-6cad49e11cb1c2a2c0ee | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB08846 |
|---|
| HMDB ID | HMDB0002899 |
|---|
| FooDB ID | FDB012575 |
|---|
| Phenol Explorer ID | 417 |
|---|
| KNApSAcK ID | C00011153 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | 3430 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Ellagic_acid |
|---|
| Chemspider ID | 4445149 |
|---|
| ChEBI ID | 4775 |
|---|
| PubChem Compound ID | 5281855 |
|---|
| Kegg Compound ID | C10788 |
|---|
| YMDB ID | YMDB01679 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=10445164 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=10999626 | | 3. https://www.ncbi.nlm.nih.gov/pubmed/?term=11902978 | | 4. https://www.ncbi.nlm.nih.gov/pubmed/?term=15659385 | | 5. https://www.ncbi.nlm.nih.gov/pubmed/?term=15936648 | | 6. https://www.ncbi.nlm.nih.gov/pubmed/?term=16317787 | | 7. https://www.ncbi.nlm.nih.gov/pubmed/?term=17379263 | | 8. https://www.ncbi.nlm.nih.gov/pubmed/?term=17940513 | | 9. https://www.ncbi.nlm.nih.gov/pubmed/?term=18155344 | | 10. https://www.ncbi.nlm.nih.gov/pubmed/?term=19015354 | | 11. https://www.ncbi.nlm.nih.gov/pubmed/?term=19684079 | | 12. https://www.ncbi.nlm.nih.gov/pubmed/?term=20034460 | | 13. https://www.ncbi.nlm.nih.gov/pubmed/?term=21922312 | | 14. https://www.ncbi.nlm.nih.gov/pubmed/?term=22173652 | | 15. https://www.ncbi.nlm.nih.gov/pubmed/?term=22538930 | | 16. https://www.ncbi.nlm.nih.gov/pubmed/?term=22626975 | | 17. https://www.ncbi.nlm.nih.gov/pubmed/?term=23058930 | | 18. https://www.ncbi.nlm.nih.gov/pubmed/?term=23060566 | | 19. https://www.ncbi.nlm.nih.gov/pubmed/?term=23092326 | | 20. https://www.ncbi.nlm.nih.gov/pubmed/?term=23322372 | | 21. https://www.ncbi.nlm.nih.gov/pubmed/?term=25438234 | | 22. https://www.ncbi.nlm.nih.gov/pubmed/?term=7644381 | | 23. Glover D, Warner ED: The CLUE test. A multiparameter coagulation and fibrinolysis screening test using the platelet aggregometer. Am J Clin Pathol. 1975 Jan;63(1):74-80. | | 24. Banzouzi JT, Prado R, Menan H, Valentin A, Roumestan C, Mallie M, Pelissier Y, Blache Y: In vitro antiplasmodial activity of extracts of Alchornea cordifolia and identification of an active constituent: ellagic acid. J Ethnopharmacol. 2002 Aug;81(3):399-401. | | 25. Cerda B, Espin JC, Parra S, Martinez P, Tomas-Barberan FA: The potent in vitro antioxidant ellagitannins from pomegranate juice are metabolised into bioavailable but poor antioxidant hydroxy-6H-dibenzopyran-6-one derivatives by the colonic microflora of healthy humans. Eur J Nutr. 2004 Aug;43(4):205-20. Epub 2004 Jan 6. | | 26. Kaiser B, Fareed J, Hoppensteadt D, Birdsong B, Walenga JM, Markwardt F: Influence of recombinant hirudin and unfractionated heparin on thrombin and factor Xa generation in extrinsic and intrinsic activated systems. Thromb Res. 1992 Jan 15;65(2):157-64. | | 27. Jimenez F, Mitts TF, Liu K, Wang Y, Hinek A: Ellagic and tannic acids protect newly synthesized elastic fibers from premature enzymatic degradation in dermal fibroblast cultures. J Invest Dermatol. 2006 Jun;126(6):1272-80. | | 28. Stoner GD, Sardo C, Apseloff G, Mullet D, Wargo W, Pound V, Singh A, Sanders J, Aziz R, Casto B, Sun X: Pharmacokinetics of anthocyanins and ellagic acid in healthy volunteers fed freeze-dried black raspberries daily for 7 days. J Clin Pharmacol. 2005 Oct;45(10):1153-64. |
|
|---|