| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 05:01:28 UTC |
|---|
| Update Date | 2016-10-28 10:03:29 UTC |
|---|
| Accession Number | CHEM015742 |
|---|
| Identification |
|---|
| Common Name | Octanoin, tri- (Octanoic acid, 1,2,3-propanetriyl ester; Tricaprylin) |
|---|
| Class | Small Molecule |
|---|
| Description | A triglyceride obtained by acylation of the three hydroxy groups of glycerol by octanoic acid. Used as an alternative energy source to glucose for patients with mild to moderate Alzheimer's disease. |
|---|
| Contaminant Sources | - Cosmetic Chemicals
- FooDB Chemicals
- OECD HPV Chemicals
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
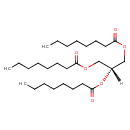
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2,3-Propanetriol trioctanoate | ChEBI | | 1,2,3-Trioctanoylglycerol | ChEBI | | Axona | ChEBI | | Caprylic acid triglyceride | ChEBI | | Caprylic acid, 1,2,3-propanetriyl ester | ChEBI | | Caprylic triglyceride | ChEBI | | Caprylin | ChEBI | | Glycerin tricaprylate | ChEBI | | Glycerol tricaprylate | ChEBI | | Glycerol trioctanoate | ChEBI | | Glycerol trioctanoin | ChEBI | | Glyceryl tricaprylate | ChEBI | | Octanoic acid triglyceride | ChEBI | | Octanoic acid, 1,1',1''-(1,2,3-propanetriyl) ester | ChEBI | | Octanoic acid, 1,2,3-propanetriyl ester | ChEBI | | Tricaprilin | ChEBI | | Tricaprylic glyceride | ChEBI | | Tricaprylin | ChEBI | | Tricapryloylglycerol | ChEBI | | Tricaprylyl glycerin | ChEBI | | Trioctanoylglyceride | ChEBI | | Trioctanoylglycerol | ChEBI | | 1,2,3-Propanetriol trioctanoic acid | Generator | | Caprylate triglyceride | Generator | | Caprylate, 1,2,3-propanetriyl ester | Generator | | Glycerin tricaprylic acid | Generator | | Glycerol tricaprylic acid | Generator | | Glycerol trioctanoic acid | Generator | | Glyceryl tricaprylic acid | Generator | | Octanoate triglyceride | Generator | | Octanoate, 1,1',1''-(1,2,3-propanetriyl) ester | Generator | | Octanoate, 1,2,3-propanetriyl ester | Generator | | 2-Ethylhexanoic acid, 1,2,3-propanetriyl ester | MeSH | | Panasate 800 | MeSH | | Glyceryl trioctanoate | MeSH | | Octanoic acid, 1,2,3- propanetriyl ester | MeSH | | Triethylhexanoin | MeSH | | Trioctanoin | MeSH | | 1-capryloyl-2-capryloyl-3-capryloyl-glycerol | Lipid Annotator, HMDB | | TAG(8:0/8:0/8:0) | Lipid Annotator, HMDB | | TAG(24:0) | Lipid Annotator, HMDB | | Tracylglycerol(24:0) | Lipid Annotator, HMDB | | Triacylglycerol | Lipid Annotator, HMDB | | Triglyceride | Lipid Annotator, HMDB | | TG(24:0) | Lipid Annotator, HMDB | | 1-octanoyl-2-octanoyl-3-octanoyl-glycerol | Lipid Annotator, HMDB | | Tracylglycerol(8:0/8:0/8:0) | Lipid Annotator, HMDB | | TG(8:0/8:0/8:0) | Lipid Annotator, ChEBI |
|
|---|
| Chemical Formula | C27H50O6 |
|---|
| Average Molecular Mass | 470.682 g/mol |
|---|
| Monoisotopic Mass | 470.361 g/mol |
|---|
| CAS Registry Number | 538-23-8 |
|---|
| IUPAC Name | 1,3-bis(octanoyloxy)propan-2-yl octanoate |
|---|
| Traditional Name | Rato |
|---|
| SMILES | [H]C(COC(=O)CCCCCCC)(COC(=O)CCCCCCC)OC(=O)CCCCCCC |
|---|
| InChI Identifier | InChI=1S/C27H50O6/c1-4-7-10-13-16-19-25(28)31-22-24(33-27(30)21-18-15-12-9-6-3)23-32-26(29)20-17-14-11-8-5-2/h24H,4-23H2,1-3H3 |
|---|
| InChI Key | VLPFTAMPNXLGLX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Triradylcglycerols |
|---|
| Direct Parent | Triacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triacyl-sn-glycerol
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Fatty acyl
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900000-2ec19e7d19282502723b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000900000-2ec19e7d19282502723b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00b9-0009700000-74eec0b6f556539a1960 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900000-b234d06f8a74d84a1ec8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000900000-b234d06f8a74d84a1ec8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00b9-0109700000-95dc0351f8ea2f8fc008 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0000900000-ed09fdff10743d8648cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0000900000-ed09fdff10743d8648cd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uji-0909900000-7a2f04eb34118f1dfedd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00fr-0205900000-ebdc13d71ea6e7ed7fb3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ufs-3988600000-ab2ef16f12938610e275 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ugj-5942000000-025420f6cf19090c67b7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000900000-65c575fb46b6dd8d287c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0000900000-65c575fb46b6dd8d287c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-0000900000-65c575fb46b6dd8d287c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00mo-0916400000-ec4e89b54212438d375c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0096-0901000000-d3d63ccae972b08be42a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0900000000-96575af340fc74287e24 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB12176 |
|---|
| HMDB ID | HMDB0011187 |
|---|
| FooDB ID | FDB003135 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Axona |
|---|
| Chemspider ID | 10393 |
|---|
| ChEBI ID | 76978 |
|---|
| PubChem Compound ID | 10850 |
|---|
| Kegg Compound ID | C13044 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=100603 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=21262136 | | 3. https://www.ncbi.nlm.nih.gov/pubmed/?term=24169853 | | 4. https://www.ncbi.nlm.nih.gov/pubmed/?term=3752242 | | 5. https://www.ncbi.nlm.nih.gov/pubmed/?term=6616756 | | 6. https://www.ncbi.nlm.nih.gov/pubmed/?term=884170 | | 7. Fendri A, Louati H, Sellami M, Gargouri H, Smichi N, Zarai Z, Aissa I, Miled N, Gargouri Y: A thermoactive uropygial esterase from chicken: purification, characterisation and synthesis of flavour esters. Int J Biol Macromol. 2012 Jun 1;50(5):1238-44. doi: 10.1016/j.ijbiomac.2012.04.008. Epub 2012 Apr 13. | | 8. Messaoudi A, Belguith H, Ben Hamida J: Three-Dimensional Structure of Arabidopsis thaliana Lipase Predicted by Homology Modeling Method. Evol Bioinform Online. 2011;7:99-105. doi: 10.4137/EBO.S7122. Epub 2011 Jun 28. | | 9. Tetrick MA, Greer FR, Benevenga NJ: Blood D-(-)-3-hydroxybutyrate concentrations after oral administration of trioctanoin, trinonanoin, or tridecanoin to newborn rhesus monkeys (Macaca mulatta). Comp Med. 2010 Dec;60(6):486-90. | | 10. Mateos-Diaz E, Rodriguez JA, de Los Angeles Camacho-Ruiz M, Mateos-Diaz JC: High-throughput screening method for lipases/esterases. Methods Mol Biol. 2012;861:89-100. doi: 10.1007/978-1-61779-600-5_5. | | 11. Ghosh S, Strum JC, Bell RM: Lipid biochemistry: functions of glycerolipids and sphingolipids in cellular signaling. FASEB J. 1997 Jan;11(1):45-50. | | 12. Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. | | 13. Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. | | 14. Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. | | 15. Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. | | 16. The lipid handbook with CD-ROM | | 17. Triglycerides and Cholesterol Research |
|
|---|