| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:29:42 UTC |
|---|
| Update Date | 2016-10-28 10:04:26 UTC |
|---|
| Accession Number | CHEM007738 |
|---|
| Identification |
|---|
| Common Name | 2,6,6-TRIMETHYLCYCLOHEX-2-ENE-1,4-DIONE |
|---|
| Class | Small Molecule |
|---|
| Description | 2,6,6-Trimethyl-2-cyclohexene-1,4-dione is found in herbs and spices. 2,6,6-Trimethyl-2-cyclohexene-1,4-dione is present in saffron (Crocus sativus) and tea; flavouring ingredient. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- STOFF IDENT Compounds
- Suspected Compounds – Schymanski Project
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
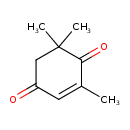
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,2,6-Trimethyl-2-cyclohexene-1,4-dione (cetoisophorone) | HMDB | | 2,2,6-Trimethylcyclohex-2-en-1,4-dione (4-oxo-isophorone) | HMDB | | 2,6,6-Trimethy-2-cyclohexene-1,4-dione (4-oxoisophorone) | HMDB | | 2,6,6-Trimethyl-2-cyclohexen-1,4-dione | HMDB | | 2,6,6-Trimethylcyclohex-2-ene-1,4-dione | HMDB | | 2-Cyclohexen-1,4-dione, 2,6,6-trimethyl | HMDB | | 3,5,5-Trimethyl-2-cyclohexene-1,4-dione | HMDB | | 3,5,5-Trimethylcyclohex-2-en-1,4-dione | HMDB | | 3,5,5-Trimethylcyclohex-2-ene-1,4-dione | HMDB | | 4-Ketoisophorone | HMDB | | 4-oxo-alpha-Isophorone | HMDB | | 4-Oxoisophorone | HMDB | | 6-Oxoisophorone | HMDB | | FEMA 3421 | HMDB | | Keto-isophorone | HMDB | | Ketoisophorone | HMDB | | Oxoisophorone | HMDB | | Oxopholone | HMDB | | Oxophorone | HMDB |
|
|---|
| Chemical Formula | C9H12O2 |
|---|
| Average Molecular Mass | 152.190 g/mol |
|---|
| Monoisotopic Mass | 152.084 g/mol |
|---|
| CAS Registry Number | 1125-21-9 |
|---|
| IUPAC Name | 2,6,6-trimethylcyclohex-2-ene-1,4-dione |
|---|
| Traditional Name | 2,6,6-trimethylcyclohex-2-ene-1,4-dione |
|---|
| SMILES | CC1=CC(=O)CC(C)(C)C1=O |
|---|
| InChI Identifier | InChI=1S/C9H12O2/c1-6-4-7(10)5-9(2,3)8(6)11/h4H,5H2,1-3H3 |
|---|
| InChI Key | AYJXHIDNNLJQDT-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cyclohexenones. Cyclohexenones are compounds containing a cylohexenone moiety, which is a six-membered aliphatic ring that carries a ketone and has one endocyclic double bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Cyclohexenones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cyclohexenone
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pbc-9400000000-ac4a8e5a500a122e8961 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1900000000-a2ede87c0b92081444bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-5900000000-43128d887aa49e04c996 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-8359b62696f36de8253d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-4d79e09516d33207e352 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1900000000-ebd44ede68a343ed2c8c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052r-9300000000-edfbab726fbefc90241c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-3cd265ceef2fb5f5db20 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-2900000000-031e7bfb242b17e4a495 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uyl-9200000000-9950f21d73347619ba59 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1900000000-bde85b5db3f5d24f0db4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-053g-9300000000-e49845f95178fde26c7e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-506b290021f45d0865df | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| | Status | Value | Unit | Sample Location | Reference |
|---|
|
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0040216 |
|---|
| FooDB ID | FDB004389 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00035002 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 56162 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 62374 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | YMDB15955 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Jerkovic I, Mastelic J, Marijanovic Z: A variety of volatile compounds as markers in unifloral honey from dalmatian sage (Salvia officinalis L.). Chem Biodivers. 2006 Dec;3(12):1307-16. | | 2. Lechtenberg M, Schepmann D, Niehues M, Hellenbrand N, Wunsch B, Hensel A: Quality and functionality of saffron: quality control, species assortment and affinity of extract and isolated saffron compounds to NMDA and sigma1 (sigma-1) receptors. Planta Med. 2008 Jun;74(7):764-72. doi: 10.1055/s-2008-1074535. Epub 2008 May 21. | | 3. Raimondi S, Roncaglia L, Amaretti A, Leonardi A, Buzzini P, Forti L, Rossi M: Rapid method for screening enoate reductase activity in yeasts. J Microbiol Methods. 2010 Nov;83(2):106-10. doi: 10.1016/j.mimet.2010.09.007. Epub 2010 Sep 16. | | 4. Goretti M, Ponzoni C, Caselli E, Marchegiani E, Cramarossa MR, Turchetti B, Forti L, Buzzini P: Bioreduction of alpha,beta-unsaturated ketones and aldehydes by non-conventional yeast (NCY) whole-cells. Bioresour Technol. 2011 Mar;102(5):3993-8. doi: 10.1016/j.biortech.2010.12.062. Epub 2010 Dec 22. | | 5. Zhuang X, Klingeman WE, Hu J, Chen F: Emission of volatile chemicals from flowering dogwood (cornus Florida L.) flowers. J Agric Food Chem. 2008 Oct 22;56(20):9570-4. doi: 10.1021/jf801651v. Epub 2008 Sep 24. | | 6. Jerkovic I, Tuberoso CI, Gugic M, Bubalo D: Composition of sulla (Hedysarum coronarium L.) honey solvent extractives determined by GC/MS: norisoprenoids and other volatile organic compounds. Molecules. 2010 Sep 9;15(9):6375-85. doi: 10.3390/molecules15096375. | | 7. Kataoka M, Kotaka A, Hasegawa A, Wada M, Yoshizumi A, Nakamori S, Shimizu S: Old Yellow Enzyme from Candida macedoniensis catalyzes the stereospecific reduction of the C=C bond of ketoisophorone. Biosci Biotechnol Biochem. 2002 Dec;66(12):2651-7. | | 8. Chen K, Sun Y, Wang C, Yao J, Chen Z, Li H: Aerobic oxidation of beta-isophorone catalyzed by N-hydroxyphthalimide: the key features and mechanism elucidated. Phys Chem Chem Phys. 2012 Sep 21;14(35):12141-6. doi: 10.1039/c2cp41617d. Epub 2012 Jul 31. | | 9. Kataoka M, Kotaka A, Thiwthong R, Wada M, Nakamori S, Shimizu S: Cloning and overexpression of the old yellow enzyme gene of Candida macedoniensis, and its application to the production of a chiral compound. J Biotechnol. 2004 Oct 19;114(1-2):1-9. | | 10. Raimondi S, Romano D, Amaretti A, Molinari F, Rossi M: Enoate reductases from non conventional yeasts: bioconversion, cloning, and functional expression in Saccharomyces cerevisiae. J Biotechnol. 2011 Dec 20;156(4):279-85. doi: 10.1016/j.jbiotec.2011.08.033. Epub 2011 Sep 16. | | 11. Hegazy ME, Hirata T, Abdel-Lateff A, el-Razek MH, Mohamed Ael-H, Hassan NM, Pare PW, Mahmoud AA: Ketoisophorone transformation by Marchantia polymorpha and Nicotiana tabacum cultured cells. Z Naturforsch C. 2008 May-Jun;63(5-6):403-8. | | 12. Mohr S, Fisher K, Scrutton NS, Goddard NJ, Fielden PR: Continuous two-phase flow miniaturised bioreactor for monitoring anaerobic biocatalysis by pentaerythritol tetranitrate reductase. Lab Chip. 2010 Aug 7;10(15):1929-36. doi: 10.1039/c003561k. Epub 2010 Jun 7. | | 13. Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC. |
|
|---|