| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:28:25 UTC |
|---|
| Update Date | 2016-11-09 01:13:38 UTC |
|---|
| Accession Number | CHEM007636 |
|---|
| Identification |
|---|
| Common Name | 1,2,5,6-TETRAHYDROCUMINIC ACID |
|---|
| Class | Small Molecule |
|---|
| Description | xi-4-Isopropyl-3-cyclohexene-1-carboxylic acid is found in herbs and spices. xi-4-Isopropyl-3-cyclohexene-1-carboxylic acid is a flavouring ingredient. xi-4-Isopropyl-3-cyclohexene-1-carboxylic acid is present in cumin oil. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
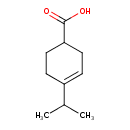
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-Isopropyl-3-cyclohexene-1-carboxylate | Generator | | 1,2,5,6-Tetrahydrocuminic acid | HMDB, MeSH | | 4-(1-Methylethyl)-3-cyclohexene-1-carboxylic acid, 9ci | HMDB | | FEMA 3731 | HMDB | | P-Menth-3-ene-7-Oic acid | HMDB | | 4-(Propan-2-yl)cyclohex-3-ene-1-carboxylate | Generator |
|
|---|
| Chemical Formula | C10H16O2 |
|---|
| Average Molecular Mass | 168.233 g/mol |
|---|
| Monoisotopic Mass | 168.115 g/mol |
|---|
| CAS Registry Number | 56424-87-4 |
|---|
| IUPAC Name | 4-(propan-2-yl)cyclohex-3-ene-1-carboxylic acid |
|---|
| Traditional Name | 4-isopropylcyclohex-3-ene-1-carboxylic acid |
|---|
| SMILES | CC(C)C1=CCC(CC1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H16O2/c1-7(2)8-3-5-9(6-4-8)10(11)12/h3,7,9H,4-6H2,1-2H3,(H,11,12) |
|---|
| InChI Key | SVVRHSSIDPLFOP-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as menthane monoterpenoids. These are monoterpenoids with a structure based on the o-, m-, or p-menthane backbone. P-menthane consists of the cyclohexane ring with a methyl group and a (2-methyl)-propyl group at the 1 and 4 ring position, respectively. The o- and m- menthanes are much rarer, and presumably arise by alkyl migration of p-menthanes. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Menthane monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - P-menthane monoterpenoid
- Monocyclic monoterpenoid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0faj-9400000000-bc2e376a04b40bddf120 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00g0-9410000000-c7825a8f3124072cd56d | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-7895fd36ceba7fba22f1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0g4i-4900000000-cbfe45b44ca00f9b705f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-1000-9100000000-500b5e4c28549c44540c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0900000000-67b2050e74ed8e34219b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xr-0900000000-2dc8adfa8057eb4eda3a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-5900000000-e156563488a4d8116aaf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-010r-9800000000-4ed7612d9268d1388444 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000x-9300000000-2a1c21967603c71a67b3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002f-9000000000-5f1fe1f7b2602f440958 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0900000000-cb7592114e600c180aec | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00xr-0900000000-492acc54af8049864732 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05dl-9500000000-7c93c8d9ad895e58040a | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037175 |
|---|
| FooDB ID | FDB016171 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 55952 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 62117 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|