| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:24:55 UTC |
|---|
| Update Date | 2016-11-09 01:09:55 UTC |
|---|
| Accession Number | CHEM007383 |
|---|
| Identification |
|---|
| Common Name | QUEBRACHO BARK EXTRACT |
|---|
| Class | Small Molecule |
|---|
| Description | An ellagitannin with a hexahydroxydiphenoyl group bridging over the 3-O and 6-O of the glucose core. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
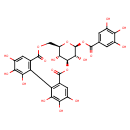
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (beta-1-O-Galloyl-3,6-(R)-hexahydroxydiphenoyl-D-glucose) | ChEBI | | 1-O-Galloyl-3,6-hexahydroxydiphenic acid-beta-D-glucopyranose | ChEBI | | (b-1-O-Galloyl-3,6-(R)-hexahydroxydiphenoyl-D-glucose) | Generator | | (Β-1-O-galloyl-3,6-(R)-hexahydroxydiphenoyl-D-glucose) | Generator | | 1-O-Galloyl-3,6-hexahydroxydiphenate-b-D-glucopyranose | Generator | | 1-O-Galloyl-3,6-hexahydroxydiphenate-beta-D-glucopyranose | Generator | | 1-O-Galloyl-3,6-hexahydroxydiphenate-β-D-glucopyranose | Generator | | 1-O-Galloyl-3,6-hexahydroxydiphenic acid-b-D-glucopyranose | Generator | | 1-O-Galloyl-3,6-hexahydroxydiphenic acid-β-D-glucopyranose | Generator | | 1-O-Galloyl-3,6-(R)-hexahydroxydiphenol-beta-D-glucose | MeSH | | Corillagin | HMDB |
|
|---|
| Chemical Formula | C27H22O18 |
|---|
| Average Molecular Mass | 634.453 g/mol |
|---|
| Monoisotopic Mass | 634.081 g/mol |
|---|
| CAS Registry Number | 977092-71-9 |
|---|
| IUPAC Name | (1S,19R,21S,22R,23R)-6,7,8,11,12,13,22,23-octahydroxy-3,16-dioxo-2,17,20-trioxatetracyclo[17.3.1.0^{4,9}.0^{10,15}]tricosa-4,6,8,10,12,14-hexaen-21-yl 3,4,5-trihydroxybenzoate |
|---|
| Traditional Name | (1S,19R,21S,22R,23R)-6,7,8,11,12,13,22,23-octahydroxy-3,16-dioxo-2,17,20-trioxatetracyclo[17.3.1.0^{4,9}.0^{10,15}]tricosa-4,6,8,10,12,14-hexaen-21-yl 3,4,5-trihydroxybenzoate |
|---|
| SMILES | O[C@@H]1[C@H]2COC(=O)C3=CC(O)=C(O)C(O)=C3C3=C(O)C(O)=C(O)C=C3C(=O)O[C@@H]1[C@@H](O)[C@H](OC(=O)C1=CC(O)=C(O)C(O)=C1)O2 |
|---|
| InChI Identifier | InChI=1S/C27H22O18/c28-9-1-6(2-10(29)16(9)32)24(39)45-27-22(38)23-19(35)13(43-27)5-42-25(40)7-3-11(30)17(33)20(36)14(7)15-8(26(41)44-23)4-12(31)18(34)21(15)37/h1-4,13,19,22-23,27-38H,5H2/t13-,19-,22-,23+,27+/m1/s1 |
|---|
| InChI Key | TUSDEZXZIZRFGC-XIGLUPEJSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydrolyzable tannins. These are tannins with a structure characterized by either of the following models. In model 1, the structure contains galloyl units (in some cases, shikimic acid units) that are linked to diverse polyol carbohydrate-, catechin-, or triterpenoid units. In model 2, contains at least two galloyl units C-C coupled to each other, and do not contain a glycosidically linked catechin unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Phenylpropanoids and polyketides |
|---|
| Class | Tannins |
|---|
| Sub Class | Hydrolyzable tannins |
|---|
| Direct Parent | Hydrolyzable tannins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydrolyzable tannin
- Macrolide
- Galloyl ester
- Gallic acid or derivatives
- P-hydroxybenzoic acid alkyl ester
- M-hydroxybenzoic acid ester
- P-hydroxybenzoic acid ester
- Benzoate ester
- Pyrogallol derivative
- Tricarboxylic acid or derivatives
- Benzenetriol
- Benzoic acid or derivatives
- Benzoyl
- 1-hydroxy-4-unsubstituted benzenoid
- Phenol
- 1-hydroxy-2-unsubstituted benzenoid
- Oxane
- Monocyclic benzene moiety
- Benzenoid
- Monosaccharide
- Lactone
- Secondary alcohol
- Carboxylic acid ester
- Acetal
- Oxacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Polyol
- Hydrocarbon derivative
- Alcohol
- Organic oxygen compound
- Organic oxide
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0kk9-1400901000-f9ee94000466cc2a5d98 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uk9-0912102000-747eb30db0e36223c7d1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0910100000-c09c146b163418500f06 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fi0-0930000000-7dda7b5a680cd0912f1b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gc0-0900214000-ac8138ca0eb7e3474411 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-1900301000-bedf60486b80a8240cc1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0lfu-4920200000-73686c08722f918b4f00 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0400709000-974c3fccc24061e5bfcb | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gbi-1900801000-39727df01a08af3c68c2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ufr-3900420000-d3d45ea63d8935f0bd13 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-016r-0900101000-19cd1c40012f2b1310f3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0i29-0900522000-f1a8081561856ad5378d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0100-5900750000-0bf5a21943a40e85788b | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0031457 |
|---|
| FooDB ID | FDB006774 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00002915 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Corilagin |
|---|
| Chemspider ID | 66248 |
|---|
| ChEBI ID | 3884 |
|---|
| PubChem Compound ID | 73568 |
|---|
| Kegg Compound ID | C10219 |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|