| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:21:02 UTC |
|---|
| Update Date | 2016-11-09 01:09:51 UTC |
|---|
| Accession Number | CHEM007064 |
|---|
| Identification |
|---|
| Common Name | PECTIN |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- HMDB Contaminants - Urine
- HPV EPA Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
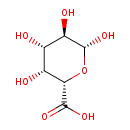
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (+)-Xylose | HMDB | | 2,3,4,5-Tetrahydroxypentanal | HMDB | | D-Lyxose | HMDB | | DL-Xylose | HMDB | | L(+)-Xylose | HMDB | | L-Lyxose | HMDB | | Lyxose | HMDB | | Pectin sugar | HMDB | | Pectinose | HMDB | | Pentose | HMDB | | Trobicin | HMDB | | Pectinic acid | HMDB | | Calcium pectinate | HMDB | | Methoxy pectin | HMDB | | Methoxylpectin | HMDB | | Methoxypectin | HMDB | | Zinc pectinate | HMDB | | Galacturonate | HMDB | | b-D-Galacturonate | HMDB | | b-D-Galacturonic acid | HMDB | | beta-D-Galacturonate | HMDB | | Β-D-galacturonate | HMDB | | Β-D-galacturonic acid | HMDB | | Pectin | MeSH | | b-D-Galactopyranuronate | Generator | | b-D-Galactopyranuronic acid | Generator | | beta-D-Galactopyranuronate | Generator | | Β-D-galactopyranuronate | Generator | | Β-D-galactopyranuronic acid | Generator |
|
|---|
| Chemical Formula | C6H10O7 |
|---|
| Average Molecular Mass | 194.139 g/mol |
|---|
| Monoisotopic Mass | 194.043 g/mol |
|---|
| CAS Registry Number | 9000-69-5 |
|---|
| IUPAC Name | (2S,3R,4S,5R,6R)-3,4,5,6-tetrahydroxyoxane-2-carboxylic acid |
|---|
| Traditional Name | β-D-galactopyranuronic acid |
|---|
| SMILES | O[C@@H]1O[C@@H]([C@H](O)[C@H](O)[C@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H10O7/c7-1-2(8)4(5(10)11)13-6(12)3(1)9/h1-4,6-9,12H,(H,10,11)/t1-,2+,3+,4-,6+/m0/s1 |
|---|
| InChI Key | AEMOLEFTQBMNLQ-DTEWXJGMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glucuronic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glucuronic acid or derivatives
- Beta-hydroxy acid
- Hydroxy acid
- Pyran
- Monosaccharide
- Oxane
- Hemiacetal
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fvi-3900000000-623cfb10797e088f31a3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-000l-6242950000-7b610ed98440b47c2a32 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00kr-6900000000-a4a0a279e2e16f432677 | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000i-9100000000-1c90dbe22f830f93ad0f | Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9000000000-cefd20d23fb5136d2104 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0900000000-142e51c19f7b152d18e6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056s-1900000000-80978d6e9cf43c2a99ae | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4r-9400000000-fe9d58fcfd8f4880243e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0007-2900000000-eecfa94a9428872c9b56 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000w-4900000000-c10f02b5bf83d62924f0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9200000000-21b195d29543b18b8595 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-2877a7e6d3ecd32cf172 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000b-7900000000-d748e59637b495442c9e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9100000000-40ac961f184de816ed86 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-3900000000-64f3d0eb26f7bc8ca624 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a73-9300000000-329488f791f643b1c308 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-474e82a5911096bc3b39 | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | Spectrum | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB03652 |
|---|
| HMDB ID | HMDB0003402 |
|---|
| FooDB ID | FDB023162 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | 34162 |
|---|
| BioCyc ID | PECTIN |
|---|
| METLIN ID | 6916 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Pectin |
|---|
| Chemspider ID | 390200 |
|---|
| ChEBI ID | 47954 |
|---|
| PubChem Compound ID | 441476 |
|---|
| Kegg Compound ID | C08348 |
|---|
| YMDB ID | YMDB00904 |
|---|
| ECMDB ID | ECMDB04097 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Klein MS, Almstetter MF, Schlamberger G, Nurnberger N, Dettmer K, Oefner PJ, Meyer HH, Wiedemann S, Gronwald W: Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J Dairy Sci. 2010 Apr;93(4):1539-50. doi: 10.3168/jds.2009-2563. | | 2. Sun HZ, Shi K, Wu XH, Xue MY, Wei ZH, Liu JX, Liu HY: Lactation-related metabolic mechanism investigated based on mammary gland metabolomics and 4 biofluids' metabolomics relationships in dairy cows. BMC Genomics. 2017 Dec 2;18(1):936. doi: 10.1186/s12864-017-4314-1. | | 3. Gu, Qu-Ming; Nickol, Robert G.; Cheng, H. N. Enzyme-catalyzed modification of pectin. Polymer Preprints (American Chemical Society, Division of Polymer Chemistry) (2003), 44(2), 608-609. | | 4. Andoh A, Bamba T, Sasaki M: Physiological and anti-inflammatory roles of dietary fiber and butyrate in intestinal functions. JPEN J Parenter Enteral Nutr. 1999 Sep-Oct;23(5 Suppl):S70-3. | | 5. Fleming SE, Marthinsen D, Kuhnlein H: Colonic function and fermentation in men consuming high fiber diets. J Nutr. 1983 Dec;113(12):2535-44. | | 6. Lewinska D, Rosinski S, Piatkiewicz W: A new pectin-based material for selective LDL-cholesterol removal. Artif Organs. 1994 Mar;18(3):217-22. | | 7. Kelsay JL, Goering HK, Behall KM, Prather ES: Effect of fiber from fruits and vegetables on metabolic responses of human subjects: fiber intakes, fecal excretions, and apparent digestibilities. Am J Clin Nutr. 1981 Sep;34(9):1849-52. | | 8. Baig MM, Cerda JJ: Pectin: its interaction with serum lipoproteins. Am J Clin Nutr. 1981 Jan;34(1):50-3. | | 9. Lairon D, Lafont H, Vigne JL, Nalbone G, Leonardi J, Hauton JC: Effects of dietary fibers and cholestyramine on the activity of pancreatic lipase in vitro. Am J Clin Nutr. 1985 Oct;42(4):629-38. | | 10. Miettinen TA, Tarpila S: Effect of pectin on serum cholesterol, fecal bile acids and biliary lipids in normolipidemic and hyperlipidemic individuals. Clin Chim Acta. 1977 Sep 1;79(2):471-7. | | 11. Bosaeus I, Carlsson NG, Sandberg AS, Andersson H: Effect of wheat bran and pectin on bile acid and cholesterol excretion in ileostomy patients. Hum Nutr Clin Nutr. 1986 Nov;40(6):429-40. | | 12. Fan TY, Feng QQ, Jia CR, Fan Q, Li CA, Bai XL: Protective effect of Weikang decoction and partial ingredients on model rat with gastric mucosa ulcer. World J Gastroenterol. 2005 Feb 28;11(8):1204-9. | | 13. Levy MC, Edwards-Levy F: Coating alginate beads with cross-linked biopolymers: a novel method based on a transacylation reaction. J Microencapsul. 1996 Mar-Apr;13(2):169-83. | | 14. Veldman FJ, Nair CH, Vorster HH, Vermaak WJ, Jerling JC, Oosthuizen W, Venter CS: Possible mechanisms through which dietary pectin influences fibrin network architecture in hypercholesterolaemic subjects. Thromb Res. 1999 Mar 15;93(6):253-64. | | 15. Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. |
|
|---|