| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:19:35 UTC |
|---|
| Update Date | 2016-11-09 01:09:50 UTC |
|---|
| Accession Number | CHEM006937 |
|---|
| Identification |
|---|
| Common Name | OCIMENE |
|---|
| Class | Small Molecule |
|---|
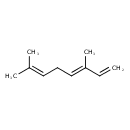
| Description | A beta-ocimene that consists of octa-1,3,6-triene bearing two methyl substituents at positions 3 and 7 (the 3E-isomer). |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (e)-3,7-Dimethylocta-1,3,6-triene | ChEBI | | 3,7-Dimethyl-1,3E,6-octatriene | ChEBI | | trans-3,7-Dimethylocta-1,3,6-triene | ChEBI | | trans-beta-Ocimene | ChEBI | | beta-Ocimene | Kegg | | trans-b-Ocimene | Generator | | trans-Β-ocimene | Generator | | b-Ocimene | Generator | | Β-ocimene | Generator | | (3E)-3,7-Dimethyl-1,3,6-octatriene | HMDB | | (3E)-3,7-Dimethylocta-1,3,6-triene | HMDB | | (e)-3,7-Dimethyloctatriene | HMDB | | (e)-beta -Ocimene | HMDB | | (E)-beta-Ocimene | HMDB | | (E)-Ocimene | HMDB | | 3,7-Dimethyl-(e)-1,3,6-octatriene | HMDB | | 3,7-Dimethyl-(e)-octatriene | HMDB | | beta -(e)-Ocimene | HMDB | | beta -trans-Ocimene | HMDB | | beta-trans-Ocimene | HMDB | | e-3,7-Dimethyl-1,3,6-octatriene | HMDB | | e-beta-Ocimene | HMDB | | trans-3,7-Dimethyl-1,3,6-octatriene | HMDB | | trans-beta -Ocimene | HMDB | | trans-Ocimene | HMDB | | (e)-b-Ocimene | Generator | | (E)-β-Ocimene | Generator | | (3E)-Ocimene | PhytoBank | | (E)-3,7-Dimethyl-1,3,6-octatriene | PhytoBank | | beta-(E)-Ocimene | PhytoBank | | β-(E)-Ocimene | PhytoBank | | β-trans-Ocimene | PhytoBank | | 3,7-Dimethyl-1,3,6-octatriene | PhytoBank |
|
|---|
| Chemical Formula | C10H16 |
|---|
| Average Molecular Mass | 136.234 g/mol |
|---|
| Monoisotopic Mass | 136.125 g/mol |
|---|
| CAS Registry Number | 13877-91-3 |
|---|
| IUPAC Name | (3E)-3,7-dimethylocta-1,3,6-triene |
|---|
| Traditional Name | β-ocimene |
|---|
| SMILES | CC(C)=CC\C=C(/C)C=C |
|---|
| InChI Identifier | InChI=1S/C10H16/c1-5-10(4)8-6-7-9(2)3/h5,7-8H,1,6H2,2-4H3/b10-8+ |
|---|
| InChI Key | IHPKGUQCSIINRJ-CSKARUKUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyclic monoterpenoids. These are monoterpenes that do not contain a cycle. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Acyclic monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acyclic monoterpenoid
- Branched unsaturated hydrocarbon
- Alkatriene
- Unsaturated aliphatic hydrocarbon
- Unsaturated hydrocarbon
- Olefin
- Acyclic olefin
- Hydrocarbon
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-Q (Non-derivatized) | splash10-0006-9100000000-3e1ffc9c9e0b47e1d6dd | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052f-9200000000-ee73cf7342b73141f4b2 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-4900000000-2eaaf884de00c6502b64 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f80-9400000000-4cc164b437465a5878ea | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-7a6775225dd2fcad42fc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0900000000-0149d94bf066009726a1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-1900000000-6c29dab24724f8b32ece | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gb9-9600000000-6b5f470455096efc86c3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-1900000000-83e72aa15e384587ebd8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-1900000000-63f5dc5fee3a530dc16f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-9000000000-cc6cf03fdb8fe15d9ed0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-3629c08e44aaa74c273c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-9000000000-6dbcb07de1e825e783ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fvi-9000000000-a9f21d2f141933dc84a1 | Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0006-9100000000-c9534182c6423a8162dc | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030089 |
|---|
| FooDB ID | FDB001465 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | C00000862 |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | CPD-4889 |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4444881 |
|---|
| ChEBI ID | 64280 |
|---|
| PubChem Compound ID | 5281553 |
|---|
| Kegg Compound ID | C09873 |
|---|
| YMDB ID | YMDB15906 |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. https://www.ncbi.nlm.nih.gov/pubmed/?term=12428002 | | 2. https://www.ncbi.nlm.nih.gov/pubmed/?term=12624761 | | 3. https://www.ncbi.nlm.nih.gov/pubmed/?term=17260255 | | 4. https://www.ncbi.nlm.nih.gov/pubmed/?term=17334921 | | 5. https://www.ncbi.nlm.nih.gov/pubmed/?term=19347799 | | 6. https://www.ncbi.nlm.nih.gov/pubmed/?term=19538549 | | 7. https://www.ncbi.nlm.nih.gov/pubmed/?term=19634337 | | 8. https://www.ncbi.nlm.nih.gov/pubmed/?term=19634338 | | 9. https://www.ncbi.nlm.nih.gov/pubmed/?term=20378980 | | 10. https://www.ncbi.nlm.nih.gov/pubmed/?term=20575390 | | 11. https://www.ncbi.nlm.nih.gov/pubmed/?term=20839643 | | 12. https://www.ncbi.nlm.nih.gov/pubmed/?term=21213991 | | 13. https://www.ncbi.nlm.nih.gov/pubmed/?term=21425693 | | 14. https://www.ncbi.nlm.nih.gov/pubmed/?term=21815435 | | 15. https://www.ncbi.nlm.nih.gov/pubmed/?term=22224304 | | 16. Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. | | 17. Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. | | 18. Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. | | 19. Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. | | 20. Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, Saito K, Kanaya S. (2012) KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012 Feb;53(2):e1. | | 21. The lipid handbook with CD-ROM |
|
|---|