| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:18:47 UTC |
|---|
| Update Date | 2016-11-09 01:09:49 UTC |
|---|
| Accession Number | CHEM006875 |
|---|
| Identification |
|---|
| Common Name | NERYL ISOBUTYRATE |
|---|
| Class | Small Molecule |
|---|
| Description | Neryl isobutyrate is a flavouring ingredient. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
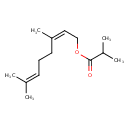
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Geranyl 2-methylpropanoic acid | Generator | | (2Z)-3,7-Dimethyl-2,6-octadienyl 2-methylpropanoate | HMDB | | (e)-3,7-Dimethyl-2,6-octadienyl 2-methylpropanoate | HMDB | | (e)-3,7-Dimethyl-2,6-octadienyl isobutyrate | HMDB | | 3,7-Dimethyl-2,6-octadienyl estertrans-isobutyric acid | HMDB | | 3,7-Dimethyl-2,6-octadienyl isobutyrate | HMDB | | 3,7-Dimethyl-isobutyratetrans-2,6-octadien-1-ol | HMDB | | FEMA 2513 | HMDB | | Geranyl isobutanoate | HMDB | | Geranyl isobutyrate | HMDB | | Isobutyric acid, (e)-3,7-dimethyl-2,6-octadienyl ester | HMDB | | Neryl iso-butyrate | HMDB | | trans-3,7-Dimethyl-2,6-octadien-1-yl 2-methylpropanoate | HMDB | | trans-3,7-Dimethyl-2,6-octadien-1-yl isobutyrate | HMDB | | trans-3-7-Dimethyl-2,6-octadienyl isobutyrate | HMDB |
|
|---|
| Chemical Formula | C14H24O2 |
|---|
| Average Molecular Mass | 224.339 g/mol |
|---|
| Monoisotopic Mass | 224.178 g/mol |
|---|
| CAS Registry Number | 2345-24-6 |
|---|
| IUPAC Name | (2Z)-3,7-dimethylocta-2,6-dien-1-yl 2-methylpropanoate |
|---|
| Traditional Name | (2Z)-3,7-dimethylocta-2,6-dien-1-yl 2-methylpropanoate |
|---|
| SMILES | CC(C)C(=O)OC\C=C(\C)CCC=C(C)C |
|---|
| InChI Identifier | InChI=1S/C14H24O2/c1-11(2)7-6-8-13(5)9-10-16-14(15)12(3)4/h7,9,12H,6,8,10H2,1-5H3/b13-9- |
|---|
| InChI Key | OGJYXQFXLSCKTP-LCYFTJDESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty alcohol esters. These are ester derivatives of a fatty alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohol esters |
|---|
| Direct Parent | Fatty alcohol esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty alcohol ester
- Monoterpenoid
- Acyclic monoterpenoid
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00kf-9000000000-0700f13d233209c71240 | Spectrum | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00kf-9000000000-0700f13d233209c71240 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-060c-9600000000-7f06f0c8f704297a2252 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004r-5960000000-c502ff66d158fef06e10 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0079-9700000000-0d017f45ec9d2963c408 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0l6u-9100000000-bf77b3a738104af1a749 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-4390000000-7a2e101210236d3bfe52 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9210000000-c47be3ffdaeface126fa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kr-9300000000-fc4e4d815d184495937d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9500000000-bc8437cb303f27665e14 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9100000000-7db5669238fb2bc13f4f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-9000000000-2f21f004481ea0ee4641 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9120000000-27970c1e919f3dd3bce4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-bccbee48d9a9e42cb5a1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0079-9000000000-8b9184596e574eb4a17c | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038260 |
|---|
| FooDB ID | FDB017554 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4517923 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 5365991 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|