| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:18:45 UTC |
|---|
| Update Date | 2016-11-09 01:09:49 UTC |
|---|
| Accession Number | CHEM006873 |
|---|
| Identification |
|---|
| Common Name | NERYL BUTYRATE |
|---|
| Class | Small Molecule |
|---|
| Description | Geranyl butyrate is found in citrus. Geranyl butyrate is found in citrus peel oils, kumquat peel oil, celery leaves/stalks, tomato, yellow passion fruit, lavender oil and other essential oils. Geranyl butyrate is a flavouring agent. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
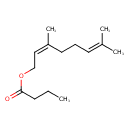
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Neryl butyric acid | Generator | | (Z)-3,7-Dimethyl-2,6-octadienyl butanoate | HMDB | | (Z)-3,7-Dimethyl-2,6-octadienyl butyrate | HMDB | | 3,7-Dimethyl-2,6-octadienyl ester(Z)-butanoic acid | HMDB | | 3,7-Dimethyl-2,6-octadienyl ester(Z)-butyric acid | HMDB | | 3,7-Dimethyl-2,6-octadienyl esterz)-butanoic acid | HMDB | | Butanoic acid, (2Z)-3,7-dimethyl-2,6-octadien-1-yl ester | HMDB | | Butanoic acid, (2Z)-3,7-dimethyl-2,6-octadienyl ester | HMDB | | cis-3,7-Dimethyl-2,6-octadienyl butanoate | HMDB | | FEMA 2774 | HMDB | | Neryl butanoate | HMDB | | Nonyl N-butyrate | HMDB | | (Z)-3,7-Dimethyl-2,6-octadienyl butyric acid | Generator | | Geranyl butyric acid | Generator |
|
|---|
| Chemical Formula | C14H24O2 |
|---|
| Average Molecular Mass | 224.339 g/mol |
|---|
| Monoisotopic Mass | 224.178 g/mol |
|---|
| CAS Registry Number | 999-40-6 |
|---|
| IUPAC Name | (2Z)-3,7-dimethylocta-2,6-dien-1-yl butanoate |
|---|
| Traditional Name | (2Z)-3,7-dimethylocta-2,6-dien-1-yl butanoate |
|---|
| SMILES | CCCC(=O)OC\C=C(\C)CCC=C(C)C |
|---|
| InChI Identifier | InChI=1S/C14H24O2/c1-5-7-14(15)16-11-10-13(4)9-6-8-12(2)3/h8,10H,5-7,9,11H2,1-4H3/b13-10- |
|---|
| InChI Key | ZSBOMYJPSRFZAL-RAXLEYEMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty alcohol esters. These are ester derivatives of a fatty alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty alcohol esters |
|---|
| Direct Parent | Fatty alcohol esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty alcohol ester
- Monoterpenoid
- Acyclic monoterpenoid
- Fatty acid ester
- Carboxylic acid ester
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014l-9100000000-0bc04e3a6c520f0cfadd | Spectrum | | GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014l-9100000000-0bc04e3a6c520f0cfadd | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-060c-9700000000-f17b16d0d76dc338c089 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004r-5960000000-5b753b26564fadc84dd6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0079-9600000000-9746fc341340ae0eca4f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gbc-9100000000-40e406f728e794d88c52 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00y0-9470000000-302dfcd1a756ed78a4d9 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kr-9200000000-50eeec28209003ce0bdd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00ko-9300000000-a79607bd760573fee44b | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9400000000-b75572ea773801a9bb22 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9100000000-8e70f8f7070d443f5535 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-1ee9f7115b0d598da89a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00dr-8290000000-194292342db985cc328a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9100000000-b6a318fb443d044e2e0e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-9000000000-9f2867696f737c8488c7 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0038259 |
|---|
| FooDB ID | FDB018005 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 4509113 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 5352162 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|