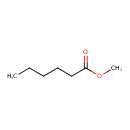
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-059f-9000000000-5fd7dd35ac31db3ce698 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00di-9000000000-1c6d18f72b4323dcbb3d | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dr-9000000000-6225c83416702a604da1 | Spectrum |
| GC-MS | GC-MS Spectrum - CI-B (Non-derivatized) | splash10-001i-3900000000-0c44c168a189853c4cf6 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dl-9000000000-8b4e4b7f45f5daa26918 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dl-9000000000-a2223723dd919d58ae77 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-059f-9000000000-5fd7dd35ac31db3ce698 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00di-9000000000-1c6d18f72b4323dcbb3d | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dr-9000000000-6225c83416702a604da1 | Spectrum |
| GC-MS | GC-MS Spectrum - CI-B (Non-derivatized) | splash10-001i-3900000000-0c44c168a189853c4cf6 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dl-9000000000-8b4e4b7f45f5daa26918 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-00dl-9000000000-a2223723dd919d58ae77 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-054o-9100000000-5c5441910ed6871e126a | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-9700000000-20aff68cecf4b94eb69d | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-053s-9300000000-2abfc6def199d6c034d0 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-a14024b94794555f817a | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-4900000000-fbf7350bd3959c6aa41c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9700000000-8044dddb59bff9baf05c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-962e1a76813769aaa8cc | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-9100000000-81b1ddc8853975968bda | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-1642da4284db2d3c9ade | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-98e8fa5a1965a4485ed8 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-21c5ae8d52f2e0f4cd33 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9000000000-c2b6445fe17cc85089a3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-20dcedc901c3e9419445 | Spectrum |
| MS | Mass Spectrum (Electron Ionization) | splash10-00dl-9000000000-75aeb95245490ac946fd | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |