| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:12:24 UTC |
|---|
| Update Date | 2016-11-09 01:09:42 UTC |
|---|
| Accession Number | CHEM006317 |
|---|
| Identification |
|---|
| Common Name | LINALYL ANTHRANILATE |
|---|
| Class | Small Molecule |
|---|
| Description | Linalyl anthranilate is used in food flavourin |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- HMDB Contaminants - Feces
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
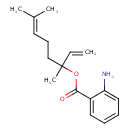
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Linalyl anthranilic acid | Generator | | 1, 6-Octadien-3-ol, 3,7-dimethyl-, 2-aminobenzoate | HMDB | | 1,5-Dimethyl-1-vinyl-4-hexen-1-yl O-aminobenzoate | HMDB | | 1,5-Dimethyl-1-vinyl-4-hexenyl anthranilate | HMDB | | 1,5-Dimethyl-1-vinylhex-4-en-1-yl 2-aminobenzoate | HMDB | | 1,6-Octadien-3-ol, 3,7-dimethyl-, 2-aminobenzoate | HMDB | | 1,6-Octadien-3-ol, 3,7-dimethyl-, 3-(2-aminobenzoate) | HMDB | | 1,6-Octadien-3-ol, 3,7-dimethyl-, O-aminobenzoate | HMDB | | 3, 7-Dimethyl-1,6-octadien-3-yl, anthranilate | HMDB | | 3,7-Dimethyl-1,6-octadien-3-ol 2-aminobenzoate | HMDB | | 3,7-Dimethyl-1,6-octadien-3-yl 2-aminobenzoate | HMDB | | 3,7-Dimethyl-1,6-octadien-3-yl O-aminobenzoate | HMDB | | 3,7-Dimethyl-1,6-octadien-3-yl, anthranilate | HMDB | | 4-Hexen-1-ol, 1,5-dimethyl-1-vinyl-, O-aminobenzoate | HMDB | | Anthranilic acid, 1, 5-dimethyl-1-vinyl-4-hexenyl ester | HMDB | | Anthranilic acid, 1,5-dimethyl-1-vinyl-4-hexenyl ester | HMDB | | Anthranilic acid, linalyl ester | HMDB | | FEMA 2637 | HMDB | | Linalyl 2-aminobenzoate | HMDB | | Linalyl O-aminobenzoate | HMDB | | 3,7-Dimethylocta-1,6-dien-3-yl 2-aminobenzoic acid | Generator |
|
|---|
| Chemical Formula | C17H23NO2 |
|---|
| Average Molecular Mass | 273.370 g/mol |
|---|
| Monoisotopic Mass | 273.173 g/mol |
|---|
| CAS Registry Number | 7149-26-0 |
|---|
| IUPAC Name | 3,7-dimethylocta-1,6-dien-3-yl 2-aminobenzoate |
|---|
| Traditional Name | 3,7-dimethylocta-1,6-dien-3-yl 2-aminobenzoate |
|---|
| SMILES | CC(C)=CCCC(C)(OC(=O)C1=CC=CC=C1N)C=C |
|---|
| InChI Identifier | InChI=1S/C17H23NO2/c1-5-17(4,12-8-9-13(2)3)20-16(19)14-10-6-7-11-15(14)18/h5-7,9-11H,1,8,12,18H2,2-4H3 |
|---|
| InChI Key | WHIJSULEEDNKPD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as aromatic monoterpenoids. These are monoterpenoids containing at least one aromatic ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Aromatic monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aminobenzoic acid or derivatives
- Aromatic monoterpenoid
- Benzoate ester
- Monocyclic monoterpenoid
- Benzoic acid or derivatives
- Aniline or substituted anilines
- Benzoyl
- Benzenoid
- Monocyclic benzene moiety
- Vinylogous amide
- Amino acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxide
- Organonitrogen compound
- Organopnictogen compound
- Amine
- Organooxygen compound
- Organic oxygen compound
- Organic nitrogen compound
- Primary amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00di-7900000000-8b372620695c2c09d114 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-1490000000-2ff0836fc2419ebb6d2f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-8930000000-e50baae1ab9a7bd24d2f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gi0-9200000000-78eff87cd6542ffe8881 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2390000000-b3cb52c5e7311404b800 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-9780000000-114d30b05b01cf86ce94 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9700000000-cc4fea34d9c8b24925bd | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-2920000000-4c5685da91926f399908 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-5900000000-de8bbb82b411d2ee1eaa | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9300000000-0a89e83f27a80cad237c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00dl-4690000000-9f23e6e36ae70f498eca | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9420000000-64dfe30866d851275fb1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-34859c653b65e95a9058 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0030433 |
|---|
| FooDB ID | FDB002297 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 22005 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 23535 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|