| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:01:47 UTC |
|---|
| Update Date | 2016-11-09 01:09:32 UTC |
|---|
| Accession Number | CHEM005404 |
|---|
| Identification |
|---|
| Common Name | 3,5-DIMETHYL-1,2-CYCLOPENTADIONE |
|---|
| Class | Small Molecule |
|---|
| Description | 3,5-Dimethyl-1,2-cyclopentanedione is found in animal foods. 3,5-Dimethyl-1,2-cyclopentanedione is a constituent of coffee and cooked cured pork. 3,5-Dimethyl-1,2-cyclopentanedione is a flavouring agent. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
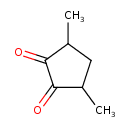
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Hydroxy-3,5-dimethyl-2-cyclopenten-1-one, 9ci | HMDB | | 3,5-Dimethyl-1,2-cyclopentadione | HMDB | | 3,5-Dimethylcyclopentane-1,2-dione | HMDB | | FEMA 3269 | HMDB | | Caramel | MeSH |
|
|---|
| Chemical Formula | C7H10O2 |
|---|
| Average Molecular Mass | 126.153 g/mol |
|---|
| Monoisotopic Mass | 126.068 g/mol |
|---|
| CAS Registry Number | 13494-07-0 |
|---|
| IUPAC Name | 3,5-dimethylcyclopentane-1,2-dione |
|---|
| Traditional Name | 3,5-dimethylcyclopentane-1,2-dione |
|---|
| SMILES | CC1CC(C)C(=O)C1=O |
|---|
| InChI Identifier | InChI=1S/C7H10O2/c1-4-3-5(2)7(9)6(4)8/h4-5H,3H2,1-2H3 |
|---|
| InChI Key | MIDXCONKKJTLDX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cyclic ketones. These are organic compounds containing a ketone that is conjugated to a cyclic moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Cyclic ketones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cyclic ketone
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-056r-9100000000-eac1e203afcdaaa9a69f | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-1900000000-31cd31914cd9466f6d9c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-8900000000-5de4f0ab6ecf92c2dc45 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-9000000000-dd57a8e6cd878b6d1ab4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-fcb80cbfcdb8990d044a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-1900000000-8ab89b622c9ad83858e1 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aor-9100000000-7e2c4ac28ab6e6fb0c95 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-1900000000-32756250330f6211ba73 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-9100000000-3a66eb05d9e211ce6bc3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aor-9100000000-c977b2b533b159464348 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-5900000000-b0ab93dccd778cb0ac92 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kf-9100000000-129a6c91b5b319814ca5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-9000000000-138d4f3db820ad834278 | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037027 |
|---|
| FooDB ID | FDB016007 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 55542 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 61634 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|