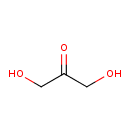
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0w2j-1900000000-b3783d91181e1657545d | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-0wa0-5900000000-f95dddc55209203877e3 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0gvk-1900000000-bd875a5207eb70c5beb8 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0w2j-1900000000-b3783d91181e1657545d | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0wa0-5900000000-f95dddc55209203877e3 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0w2j-2900000000-f84cc49dd11822409add | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a59-9000000000-089abb74f439c9265bb6 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00g0-9320000000-6bdc226e79d3bc473a66 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9000000000-7bb646a5cb5ca825e7b5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05fu-9000000000-ab60a50ef2013d762751 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-ef7c943cff3a9adfd1ed | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-0e836f7fe0512bb62662 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-059i-9000000000-d1a13699d0d0cbea36b5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-57da9a85ff22593c3ab3 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-9000000000-3deebe0c487d3443c844 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006x-9000000000-73f63f3caf2a9bf8b899 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-195717f89ffa991dcb57 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9000000000-55ab07d1079db56cca4c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ab9-9000000000-f9afe28f3ce4c2f913d5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-8e71e2b93a16e2be3db1 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | Spectrum |