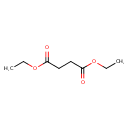
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-004i-6900000000-91b4bd8842569eab361f | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fb9-6900000000-7c630102d002ffd92fa9 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0ufr-5900000000-447fd91aad895684276e | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fb9-6900000000-940ba7631d979c2aaa49 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0ufr-3900000000-7f6aa2df583c7cd0c94b | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-004i-6900000000-91b4bd8842569eab361f | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fb9-6900000000-7c630102d002ffd92fa9 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0ufr-5900000000-447fd91aad895684276e | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0fb9-6900000000-940ba7631d979c2aaa49 | Spectrum |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0ufr-3900000000-7f6aa2df583c7cd0c94b | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fb9-9700000000-0513cf8f1fe77b0ed5af | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-9000000000-91c46e82328c3fe47ad9 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-98c4790a0a7938d84e5b | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-05fr-9100000000-64c2e21d9f9b1063a9a4 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-8900000000-666ceb07119104eed97a | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-7900000000-cefa399217e8e3a20487 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-b1f0be6edc02308e2cf0 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-1900000000-7c0b11b197a1f93dfa39 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004s-8900000000-83fc620612da3d1c067f | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0571-9100000000-b25194b84647b74001b5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00fr-2900000000-30765b159461667846d4 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0092-8900000000-cfb7037b49667188b454 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9200000000-44495c741511a36e6b58 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fb9-3900000000-4b50fb0509e11b2802c9 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0umj-9700000000-75cd91ea8a37bec260cc | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05fs-9100000000-57d3f3e93c4ab17eb6ff | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00dr-6900000000-e4d16a891fbf23cbea67 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000l-9100000000-c390ad33e07af6ecd735 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9100000000-8dcd782d35b36f7a0d1a | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |