| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 02:00:24 UTC |
|---|
| Update Date | 2016-11-09 01:09:31 UTC |
|---|
| Accession Number | CHEM005307 |
|---|
| Identification |
|---|
| Common Name | DEHYDRODIHYDROIONONE |
|---|
| Class | Small Molecule |
|---|
| Description | A member of the class of cyclohexadienes that is cyclohexa-1,3-diene substituted by a 3-oxobutyl group at position 1 and by methyl groups at positions 2 and 6. |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- HMDB Contaminants - Urine
|
|---|
| Contaminant Type | Not Available |
|---|
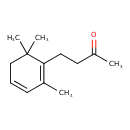
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,4-Didehydro-7,8-dihydro-beta-ionone | ChEBI | | 4-(2,6,6-Trimethyl-1,3-cyclohexadien-1-yl)butan-2-one | ChEBI | | 4-(2,6,6-Trimethyl-1,3-cyclohexadienyl)-2-butanone | ChEBI | | 4-(2,6,6-Trimethylcyclohexa-1,3-dien-1-yl)butan-2-one | ChEBI | | 4-(2,6,6-Trimethylcyclohexa-1,3-dienyl)butan-2-one | ChEBI | | 7,8-Dihydro-3,4-dehydro-beta-ionone | ChEBI | | Dehydrodihydroionone | ChEBI | | FEMA 3447 | ChEBI | | 3,4-Didehydro-7,8-dihydro-b-ionone | Generator | | 3,4-Didehydro-7,8-dihydro-β-ionone | Generator | | 7,8-Dihydro-3,4-dehydro-b-ionone | Generator | | 7,8-Dihydro-3,4-dehydro-β-ionone | Generator | | 4-(2,6,6-Trimethyl cyclohexa-1,3-dienyl) butane-2-one | HMDB | | dihydrodehydro-beta-Ionone | HMDB |
|
|---|
| Chemical Formula | C13H20O |
|---|
| Average Molecular Mass | 192.297 g/mol |
|---|
| Monoisotopic Mass | 192.151 g/mol |
|---|
| CAS Registry Number | 20483-36-7 |
|---|
| IUPAC Name | 4-(2,6,6-trimethylcyclohexa-1,3-dien-1-yl)butan-2-one |
|---|
| Traditional Name | 4-(2,6,6-trimethylcyclohexa-1,3-dien-1-yl)butan-2-one |
|---|
| SMILES | CC(=O)CCC1=C(C)C=CCC1(C)C |
|---|
| InChI Identifier | InChI=1S/C13H20O/c1-10-6-5-9-13(3,4)12(10)8-7-11(2)14/h5-6H,7-9H2,1-4H3 |
|---|
| InChI Key | SQFRYZPEWOZAKJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sesquiterpenoids. These are terpenes with three consecutive isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Megastigmane sesquiterpenoid
- Sesquiterpenoid
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002f-5900000000-5736fe8e960c46e93ba3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002f-1900000000-0e04520fc1c4c99c5426 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00p3-4900000000-95189d759958da4cd7db | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gb9-9400000000-a0c1a282f3fe2da24144 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-88505512782936b3ed7e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-2900000000-d9772c13d17a491dc87d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9600000000-d83f781d2e263e20e007 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-0900000000-25eb405119b32dd9570a | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05g3-5900000000-f9568278d81f5d6dc1b3 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05mo-9400000000-cdeeb27239a996dcb3ab | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-14436e4d9ff8d032db54 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dl-2900000000-4fc59527a3e46530a631 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052e-5900000000-68b6ab45c521a59d612e | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0037139 |
|---|
| FooDB ID | FDB016134 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 501713 |
|---|
| ChEBI ID | 88549 |
|---|
| PubChem Compound ID | 577126 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|