| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-19 01:58:30 UTC |
|---|
| Update Date | 2016-11-09 01:09:29 UTC |
|---|
| Accession Number | CHEM005135 |
|---|
| Identification |
|---|
| Common Name | CIS- AND TRANS-ETHYL 2,4-DIMETHYL-1,3-DIOXOLANE-2-ACETATE |
|---|
| Class | Small Molecule |
|---|
| Description | cis- and trans-Ethyl 2,4-dimethyl-1,3-dioxolane-2-acetate is used as a food additive [EAFUS] ("EAFUS: Everything Added to Food in the United States. [http://www.eafus.com/]") |
|---|
| Contaminant Sources | - EAFUS Chemicals
- FooDB Chemicals
- ToxCast & Tox21 Chemicals
|
|---|
| Contaminant Type | Not Available |
|---|
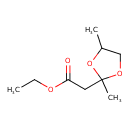
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| cis- And trans-ethyl 2,4-dimethyl-1,3-dioxolane-2-acetic acid | Generator | | 1,3-Dioxolane-2-acetic acid, 2,4-dimethyl-, ethyl ester | HMDB | | Acetoacetic acid, ethyl ester, 1,2-propylene ketal | HMDB | | Ethyl (2,4-dimethyl-1,3-dioxolan-2-yl)acetate | HMDB | | Ethyl 2, 4-dimethyl-1,3-dioxolane-2-acetate | HMDB | | Ethyl 2,4-dimethyl-1,3-dioxolane-2-acetate | HMDB | | Ethyl acetoacetate propylene glycol ketal | HMDB | | Ethyl dimethyl dioxolane acetate | HMDB |
|
|---|
| Chemical Formula | C9H16O4 |
|---|
| Average Molecular Mass | 188.221 g/mol |
|---|
| Monoisotopic Mass | 188.105 g/mol |
|---|
| CAS Registry Number | 6290-17-1 |
|---|
| IUPAC Name | ethyl 2-(2,4-dimethyl-1,3-dioxolan-2-yl)acetate |
|---|
| Traditional Name | ethyl 2-(2,4-dimethyl-1,3-dioxolan-2-yl)acetate |
|---|
| SMILES | CCOC(=O)CC1(C)OCC(C)O1 |
|---|
| InChI Identifier | InChI=1S/C9H16O4/c1-4-11-8(10)5-9(3)12-6-7(2)13-9/h7H,4-6H2,1-3H3 |
|---|
| InChI Key | GSIXJEIRJVOUFB-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acid esters. These are carboxylic ester derivatives of a fatty acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acid esters |
|---|
| Direct Parent | Fatty acid esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Ketal
- Fatty acid ester
- Meta-dioxolane
- Carboxylic acid ester
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Acetal
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ufr-7900000000-63474ed11fc6a85e1451 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1900000000-a1aea8d7e3c043dc2694 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-4900000000-c5f6870581fd024b2501 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00tg-9100000000-0824cc156e115e291813 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000l-2900000000-9d8ea61e3f3ba1722560 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kp-4900000000-1d8b7aa9667f46bf2aba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-7900000000-67446b8db35780acf915 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-2900000000-78a9085629910ce18434 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052f-9800000000-faafb1cc0dd8bcd1892f | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bt9-9400000000-04f60e8b52959429ed32 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-3900000000-74bc938b99186eaf8f96 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0hix-9800000000-8c907ff414e617b4558e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9200000000-c856aec7b6f188683f4c | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum | | 1D NMR | 1H NMR Spectrum | Not Available | Spectrum | | 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0032200 |
|---|
| FooDB ID | FDB009181 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| Chemspider ID | 86088 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 95392 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | |
|---|