| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-05-25 18:51:22 UTC |
|---|
| Update Date | 2016-11-09 01:17:34 UTC |
|---|
| Accession Number | CHEM022766 |
|---|
| Identification |
|---|
| Common Name | Tauroursodeoxycholic acid |
|---|
| Class | Small Molecule |
|---|
| Description | |
|---|
| Contaminant Sources | - FooDB Chemicals
- HMDB Contaminants - Feces
|
|---|
| Contaminant Type | Not Available |
|---|
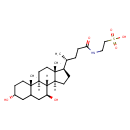
| Chemical Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Tauroursodeoxycholate | Generator | | 2-(((3-alpha,5-beta,7-beta)-3,7-Dihydroxy-24-oxocholan-24-yl) amino)ethanesulfonate | HMDB | | 2-(((3-alpha,5-beta,7-beta)-3,7-Dihydroxy-24-oxocholan-24-yl) amino)ethanesulfonic acid | HMDB | | 2-(((3-alpha,5-beta,7-beta)-3,7-Dihydroxy-24-oxocholan-24-yl)amino)-ethanesulfonate | HMDB | | 2-(((3-alpha,5-beta,7-beta)-3,7-Dihydroxy-24-oxocholan-24-yl)amino)-ethanesulfonic acid | HMDB | | 3a,7b-Dihydroxy-5b-cholanoyltaurine | HMDB | | N-(3-alpha,7-beta-Dihydroxy-5-beta-cholan-24-oyl)-taurine | HMDB | | TUDCA | HMDB | | UR 906 | HMDB | | Ursodeoxycholyltaurine | HMDB | | Tauroursodeoxycholic acid, monosodium salt, (3alpha,5beta,7alpha)-isomer | HMDB | | Tauroursodeoxycholic acid, (3alpha,5alpha,7alpha)-isomer | HMDB | | Tauroursodeoxycholic acid, monosodium salt, (3alpha,7alpha)-isomer | HMDB | | (4R)-4-[(1S,2S,5R,9S,10R,11S,14R,15R)-5,9-Dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-N-(2-sulfoethyl)pentanimidate | HMDB | | (4R)-4-[(1S,2S,5R,9S,10R,11S,14R,15R)-5,9-Dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-N-(2-sulphoethyl)pentanimidate | HMDB | | (4R)-4-[(1S,2S,5R,9S,10R,11S,14R,15R)-5,9-Dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-N-(2-sulphoethyl)pentanimidic acid | HMDB |
|
|---|
| Chemical Formula | C26H45NO6S |
|---|
| Average Molecular Mass | 499.704 g/mol |
|---|
| Monoisotopic Mass | 499.297 g/mol |
|---|
| CAS Registry Number | 14605-22-2 |
|---|
| IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,9S,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanamido]ethane-1-sulfonic acid |
|---|
| Traditional Name | 2-[(4R)-4-[(1S,2S,5R,9S,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanamido]ethanesulfonic acid |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)CCC(=O)NCCS(O)(=O)=O)[C@@]1(C)CC[C@@]1([H])[C@@]2([H])[C@@H](O)CC2C[C@H](O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C26H45NO6S/c1-16(4-7-23(30)27-12-13-34(31,32)33)19-5-6-20-24-21(9-11-26(19,20)3)25(2)10-8-18(28)14-17(25)15-22(24)29/h16-22,24,28-29H,4-15H2,1-3H3,(H,27,30)(H,31,32,33)/t16-,17?,18-,19-,20+,21+,22+,24+,25+,26-/m1/s1 |
|---|
| InChI Key | BHTRKEVKTKCXOH-VSHSPWMTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as taurinated bile acids and derivatives. These are bile acid derivatives containing a taurine conjugated to the bile acid moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Taurinated bile acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Taurinated bile acid
- Dihydroxy bile acid, alcohol, or derivatives
- Hydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- 7-alpha-hydroxysteroid
- 7-hydroxysteroid
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Sulfonyl
- Organosulfonic acid
- Organosulfonic acid or derivatives
- Organic sulfonic acid or derivatives
- Alkanesulfonic acid
- Cyclic alcohol
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Carboxylic acid derivative
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Organonitrogen compound
- Organosulfur compound
- Organooxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Not Available |
|---|
| Cellular Locations | Not Available |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Appearance | Not Available |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053r-0212900000-14bc097af3f7499753d7 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-004i-4214259000-4a72734ef61b01c2f556 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_6) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00e9-0801920000-ecfc97935416325b3ca5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-2901100000-9a3cf7f2963c041be363 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-8904100000-98cfa2606dbba4ef47a0 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000t-2001900000-4adf0e40de3812e8d0ba | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-6404900000-4d05b6f0352aebf4228d | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-9102000000-63892e6e06f1123f8bfc | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-31f17d1d48b7afdf3bae | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0000900000-f4f03ef481870394f9e2 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9100300000-654e1207ef97c7662b6e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gx0-0000940000-9827b7f4a5b2f54a7bd4 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0nmi-4639730000-a8491d87ed7e9a353ee5 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f7x-3893000000-cc5d0b26da084a4d3a35 | Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | Not Available |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB0000874 |
|---|
| FooDB ID | FDB022294 |
|---|
| Phenol Explorer ID | Not Available |
|---|
| KNApSAcK ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| METLIN ID | 5835 |
|---|
| PDB ID | Not Available |
|---|
| Wikipedia Link | Tauroursodeoxycholic acid |
|---|
| Chemspider ID | 25949321 |
|---|
| ChEBI ID | Not Available |
|---|
| PubChem Compound ID | 12443252 |
|---|
| Kegg Compound ID | Not Available |
|---|
| YMDB ID | Not Available |
|---|
| ECMDB ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | | 1. Zhuo, Chao; Feng, Wei; Wu, Da-jun; Xiong, Zhi-gang. Synthesis of tauroursodeoxycholic acid. Hecheng Huaxue (2002), 10(5), 444-446. | | 2. Rolo AP, Palmeira CM, Holy JM, Wallace KB: Role of mitochondrial dysfunction in combined bile acid-induced cytotoxicity: the switch between apoptosis and necrosis. Toxicol Sci. 2004 May;79(1):196-204. Epub 2004 Feb 19. | | 3. Tessier E, Neirinck L, Zhu Z: High-performance liquid chromatographic mass spectrometric method for the determination of ursodeoxycholic acid and its glycine and taurine conjugates in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2003 Dec 25;798(2):295-302. | | 4. Yamaguchi Y, Itami S, Nishida K, Ando Y, Okamoto S, Hosokawa K, Yoshikawa K: Taurin-conjugated ursodeoxycholic acid has a reversible inhibitory effect on human keratinocyte growth. J Dermatol Sci. 1998 Sep;18(1):35-42. | | 5. Shekels LL, Beste JE, Ho SB: Tauroursodeoxycholic acid protects in vitro models of human colonic cancer cells from cytotoxic effects of hydrophobic bile acids. J Lab Clin Med. 1996 Jan;127(1):57-66. | | 6. Lee DK, Park SY, Baik SK, Kwon SO, Chung JM, Oh ES, Kim HS: [Deoxycholic acid-induced signal transduction in HT-29 cells: role of NF-kappa B and interleukin-8]. Korean J Gastroenterol. 2004 Mar;43(3):176-85. | | 7. Cai W, Khaoustov VI, Xie Q, Pan T, Le W, Yoffe B: Interferon-alpha-induced modulation of glucocorticoid and serotonin receptors as a mechanism of depression. J Hepatol. 2005 Jun;42(6):880-7. Epub 2005 Apr 7. | | 8. Loria P, Bozzoli M, Concari M, Guicciardi ME, Carubbi F, Bertolotti M, Piani D, Nistri A, Angelico M, Romani M, Carulli N: Effect of taurohyodeoxycholic acid on biliary lipid secretion in humans. Hepatology. 1997 Jun;25(6):1306-14. | | 9. Henzel K, Thorborg C, Hofmann M, Zimmer G, Leuschner U: Toxicity of ethanol and acetaldehyde in hepatocytes treated with ursodeoxycholic or tauroursodeoxycholic acid. Biochim Biophys Acta. 2004 Feb 2;1644(1):37-45. | | 10. St-Pierre MV, Kullak-Ublick GA, Hagenbuch B, Meier PJ: Transport of bile acids in hepatic and non-hepatic tissues. J Exp Biol. 2001 May;204(Pt 10):1673-86. | | 11. Claudel T, Staels B, Kuipers F: The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005 Oct;25(10):2020-30. Epub 2005 Jul 21. | | 12. Chiang JY: Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003 Mar;284(3):G349-56. | | 13. Davis RA, Miyake JH, Hui TY, Spann NJ: Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002 Apr;43(4):533-43. |
|
|---|